Riboflavin production in Lactococcus lactis: potential for in situ production of vitamin-enriched foods
- PMID: 15466513
- PMCID: PMC522069
- DOI: 10.1128/AEM.70.10.5769-5777.2004
Riboflavin production in Lactococcus lactis: potential for in situ production of vitamin-enriched foods
Abstract
This study describes the genetic analysis of the riboflavin (vitamin B(2)) biosynthetic (rib) operon in the lactic acid bacterium Lactococcus lactis subsp. cremoris strain NZ9000. Functional analysis of the genes of the L. lactis rib operon was performed by using complementation studies, as well as by deletion analysis. In addition, gene-specific genetic engineering was used to examine which genes of the rib operon need to be overexpressed in order to effect riboflavin overproduction. Transcriptional regulation of the L. lactis riboflavin biosynthetic process was investigated by using Northern hybridization and primer extension, as well as the analysis of roseoflavin-induced riboflavin-overproducing L. lactis isolates. The latter analysis revealed the presence of both nucleotide replacements and deletions in the regulatory region of the rib operon. The results presented here are an important step toward the development of fermented foods containing increased levels of riboflavin, produced in situ, thus negating the need for vitamin fortification.
Figures




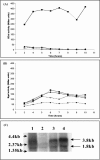
References
-
- Bacher, A., S. Eberhardt, M. Fischer, K. Kis, and G. Richter. 2000. Biosynthesis of vitamin B2 (riboflavin). Annu. Rev. Nutr. 20:153-167. - PubMed
-
- Bacher, A., S. Eberhardt, and G. Richter. 1996. Biosynthesis of riboflavin, p. 657-664. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
-
- Bandrin, S. V., P. M. Rabinovich, and A. I. Stepanov. 1983. Three linkage groups of the genes of riboflavin biosynthesis in Escherichia coli. Genetika 19:1419-1425. (In Russian.) - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

