Discrimination of modes of action of antifungal substances by use of metabolic footprinting
- PMID: 15466562
- PMCID: PMC522091
- DOI: 10.1128/AEM.70.10.6157-6165.2004
Discrimination of modes of action of antifungal substances by use of metabolic footprinting
Abstract
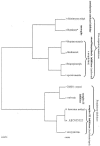
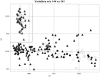
Diploid cells of Saccharomyces cerevisiae were grown under controlled conditions with a Bioscreen instrument, which permitted the essentially continuous registration of their growth via optical density measurements. Some cultures were exposed to concentrations of a number of antifungal substances with different targets or modes of action (sterol biosynthesis, respiratory chain, amino acid synthesis, and the uncoupler). Culture supernatants were taken and analyzed for their "metabolic footprints" by using direct-injection mass spectrometry. Discriminant function analysis and hierarchical cluster analysis allowed these antifungal compounds to be distinguished and classified according to their modes of action. Genetic programming, a rule-evolving machine learning strategy, allowed respiratory inhibitors to be discriminated from others by using just two masses. Metabolic footprinting thus represents a rapid, convenient, and information-rich method for classifying the modes of action of antifungal substances.
Figures




References
-
- Allen, J. K., H. M. Davey, D. Broadhurst, J. K. Heald, J. J. Rowland, S. G. Oliver, and D. B. Kell. 2003. High-throughput characterisation of yeast mutants for functional genomics using metabolic footprinting. Nat. Biotechnol. 21:692-696. - PubMed
-
- Aranibar, N., B. K. Singh, G. W. Stockton, and K.-H. Ott. 2001. Automated mode-of-action detection by metabolic profiling. Biochem. Biophys. Res. Commun. 286:150-155. - PubMed
-
- Banzhaf, W., P. Nordin, R. E. Keller, and F. D. Francone. 1998. Genetic programming: an introduction. Morgan Kaufmann, San Francisco, Calif.
-
- Castrillo, J. I., A. Hayes, S. Mohammed, S. J. Gaskell, and S. G. Oliver. 2003. An optimized protocol for metabolome analysis in yeast using direct infusion electrospray mass spectrometry. Phytochemistry 62:929-937. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

