Metabolically active eukaryotic communities in extremely acidic mine drainage
- PMID: 15466574
- PMCID: PMC522060
- DOI: 10.1128/AEM.70.10.6264-6271.2004
Metabolically active eukaryotic communities in extremely acidic mine drainage
Abstract
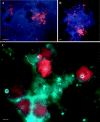
Acid mine drainage (AMD) microbial communities contain microbial eukaryotes (both fungi and protists) that confer a biofilm structure and impact the abundance of bacteria and archaea and the community composition via grazing and other mechanisms. Since prokaryotes impact iron oxidation rates and thus regulate AMD generation rates, it is important to analyze the fungal and protistan populations. We utilized 18S rRNA and beta-tubulin gene phylogenies and fluorescent rRNA-specific probes to characterize the eukaryotic diversity and distribution in extremely acidic (pHs 0.8 to 1.38), warm (30 to 50 degrees C), metal-rich (up to 269 mM Fe(2+), 16.8 mM Zn, 8.5 mM As, and 4.1 mM Cu) AMD solutions from the Richmond Mine at Iron Mountain, Calif. A Rhodophyta (red algae) lineage and organisms from the Vahlkampfiidae family were identified. The fungal 18S rRNA and tubulin gene sequences formed two distinct phylogenetic groups associated with the classes Dothideomycetes and Eurotiomycetes. Three fungal isolates that were closely related to the Dothideomycetes clones were obtained. We suggest the name "Acidomyces richmondensis" for these isolates. Since these ascomycete fungi were morphologically indistinguishable, rRNA-specific oligonucleotide probes were designed to target the Dothideomycetes and Eurotiomycetes via fluorescent in situ hybridization (FISH). FISH analyses indicated that Eurotiomycetes are generally more abundant than Dothideomycetes in all of the seven locations studied within the Richmond Mine system. This is the first study to combine the culture-independent detection of fungi with in situ detection and a demonstration of activity in an acidic environment. The results expand our understanding of the subsurface AMD microbial community structure.
Figures



References
-
- Amaral Zettler, L. A., F. Gómez, E. R. Zettler, B. G. Keenan, R. Amils, and M. L. Sogin. 2002. Eukaryotic diversity in Spain's river of fire. Nature 417:137. - PubMed
-
- Amaral Zettler, L. A., M. A. Messerli, A. D. Laatsch, P. J. S. Smith, and M. L. Sogin. 2003. From genes to genomes: beyond biodiversity in Spain's Rio Tinto. Biol. Bull. 204:205-209. - PubMed
-
- Baker, B. J., and J. F. Banfield. 2003. Microbial communities in acid mine drainage. FEMS Microbiol. Ecol. 44:139-152. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

