Identitag, a relational database for SAGE tag identification and interspecies comparison of SAGE libraries
- PMID: 15469608
- PMCID: PMC535903
- DOI: 10.1186/1471-2105-5-143
Identitag, a relational database for SAGE tag identification and interspecies comparison of SAGE libraries
Abstract
Background: Serial Analysis of Gene Expression (SAGE) is a method of large-scale gene expression analysis that has the potential to generate the full list of mRNAs present within a cell population at a given time and their frequency. An essential step in SAGE library analysis is the unambiguous assignment of each 14 bp tag to the transcript from which it was derived. This process, called tag-to-gene mapping, represents a step that has to be improved in the analysis of SAGE libraries. Indeed, the existing web sites providing correspondence between tags and transcripts do not concern all species for which numerous EST and cDNA have already been sequenced.
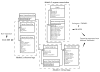
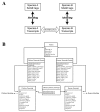
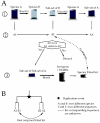
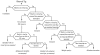
Results: This is the reason why we designed and implemented a freely available tool called Identitag for tag identification that can be used in any species for which transcript sequences are available. Identitag is based on a relational database structure in order to allow rapid and easy storage and updating of data and, most importantly, in order to be able to precisely define identification parameters. This structure can be seen like three interconnected modules : the first one stores virtual tags extracted from a given list of transcript sequences, the second stores experimental tags observed in SAGE experiments, and the third allows the annotation of the transcript sequences used for virtual tag extraction. It therefore connects an observed tag to a virtual tag and to the sequence it comes from, and then to its functional annotation when available. Databases made from different species can be connected according to orthology relationship thus allowing the comparison of SAGE libraries between species. We successfully used Identitag to identify tags from our chicken SAGE libraries and for chicken to human SAGE tags interspecies comparison. Identitag sources are freely available on http://pbil.univ-lyon1.fr/software/identitag/ web site.
Conclusions: Identitag is a flexible and powerful tool for tag identification in any single species and for interspecies comparison of SAGE libraries. It opens the way to comparative transcriptomic analysis, an emerging branch of biology.
Figures
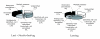
References
-
- Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. - PubMed
-
- SAGE Genie http://cgap.nci.nih.gov/SAGE
-
- Melbourne Brain Genome Project http://www.mbgproject.org/
-
- Mouse SAGE site http://mouse.biomed.cas.cz/sage/
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

