Three-body interactions improve the prediction of rate and mechanism in protein folding models
- PMID: 15469920
- PMCID: PMC524050
- DOI: 10.1073/pnas.0403486101
Three-body interactions improve the prediction of rate and mechanism in protein folding models
Abstract
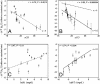
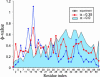
Here we study the effects of many-body interactions on rate and mechanism in protein folding by using the results of molecular dynamics simulations on numerous coarse-grained Calpha-model single-domain proteins. After adding three-body interactions explicitly as a perturbation to a Gō-like Hamiltonian with native pairwise interactions only, we have found (i) a significantly increased correlation with experimental phi values and folding rates, (ii) a stronger correlation of folding rate with contact order, matching the experimental range in rates when the fraction of three-body energy in the native state is approximately 20%, and (iii) a considerably larger amount of three-body energy present in chymotripsin inhibitor than in the other proteins studied.
Figures



References
-
- Wolynes, P. G., Onuchic, J. N. & Thirumalai, D. (1995) Science 267, 1619–1620. - PubMed
-
- Dobson, C. M., Sali, A. & Karplus, M. (1998) Angew. Chem. Int. Ed. 37, 868–893. - PubMed
-
- Fersht, A. R. (1999) Structure and Mechanism in Protein Science (Freeman, New York), 1st Ed.
-
- Mirny, L. & Shakhnovich, E. (2001) Annu. Rev. Biophys. Biomol. Struct. 30, 361–396. - PubMed
-
- Dill, K. A. & Chan, H. S. (1997) Nat. Struct. Biol. 4, 10–19. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

