Comprehensive gene expression profiles reveal pathways related to the pathogenesis of chronic obstructive pulmonary disease
- PMID: 15469929
- PMCID: PMC522001
- DOI: 10.1073/pnas.0401168101
Comprehensive gene expression profiles reveal pathways related to the pathogenesis of chronic obstructive pulmonary disease
Abstract
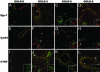
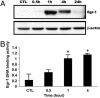
To better understand the molecular basis of chronic obstructive pulmonary disease (COPD), we used serial analysis of gene expression (SAGE) and microarray analysis to compare the gene expression patterns of lung tissues from COPD and control smokers. A total of 59,343 tags corresponding to 26,502 transcripts were sequenced in SAGE analyses. A total of 327 genes were differentially expressed (1.5-fold up- or down-regulated). Microarray analysis using the same RNA source detected 261 transcripts that were differentially expressed to a significant degree between GOLD-2 and GOLD-0 smokers. We confirmed the altered expression of a select number of genes by using real-time quantitative RT-PCR. These genes encode for transcription factors (EGR1 and FOS), growth factors or related proteins (CTGF, CYR61, CX3CL1, TGFB1, and PDGFRA), and extracellular matrix protein (COL1A1). Immunofluorescence studies on the same lung specimens localized the expression of Egr-1, CTGF, and Cyr61 to alveolar epithelial cells, airway epithelial cells, and stromal and inflammatory cells of GOLD-2 smokers. Cigarette smoke extract induced Egr-1 protein expression and increased Egr-1 DNA-binding activity in human lung fibroblast cells. Cytomix (tumor necrosis factor alpha, IL-1beta, and IFN-gamma) treatment showed that the activity of matrix metalloproteinase-2 (MMP-2) was increased in lung fibroblasts from EGR1 control (+/+) mice but not detected in that of EGR1 null (-/-) mice, whereas MMP-9 was regulated by EGR1 in a reverse manner. Our study represents the first comprehensive analysis of gene expression on GOLD-2 versus GOLD-0 smokers and reveals previously unreported candidate genes that may serve as potential molecular targets in COPD.
Figures



References
-
- Voelkel, N. F. & MacNee, W. (2002) in Chronic Obstructive Lung Diseases (BC Decker, Hamilton, ON, Canada), pp. 90-113.
-
- Petty, T. L. (2002) Chest 121, 116s-120s. - PubMed
-
- Sethi, J. M. & Rochester C. L. (2000) Clin. Chest Med. 21, 1-26. - PubMed
-
- Mayer, A. S. & Mewman, L. S. (2001) Respir. Physiol. 128, 3-11. - PubMed
-
- Pauwels, R. A., Buist, A. S., Calverley, P. M. A., Jenkins, C. R. & Jurd, S. S. (2001) Am. J. Respir. Crit. Care Med. 163, 1256-1276. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

