Developmentally regulated switch in alternatively spliced SNAP-25 isoforms alters facilitation of synaptic transmission
- PMID: 15470145
- PMCID: PMC6729955
- DOI: 10.1523/JNEUROSCI.1940-04.2004
Developmentally regulated switch in alternatively spliced SNAP-25 isoforms alters facilitation of synaptic transmission
Abstract
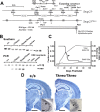
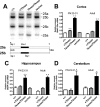
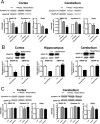
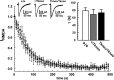
Although the basic molecular components that promote regulated neurotransmitter release are well established, the contribution of these proteins as regulators of the plasticity of neurotransmission and refinement of synaptic connectivity during development is elaborated less fully. For example, during the period of synaptic growth and maturation in brain, the expression of synaptosomal protein 25 kDa (SNAP-25), a neuronal t-SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) essential for action potential-dependent neuroexocytosis, is altered through alternative splicing of pre-mRNA transcripts. We addressed the role of the two splice-variant isoforms of SNAP-25 with a targeted mouse mutation that impairs the shift from SNAP-25a to SNAP-25b. Most of these mutant mice die between 3 and 5 weeks of age, which coincides with the time when SNAP-25b expression normally reaches mature levels in brain and synapse formation is essentially completed. The altered expression of these SNAP-25 isoforms influences short-term synaptic function by affecting facilitation but not the initial probability of release. This suggests that mechanisms controlling alternative splicing between SNAP-25 isoforms contribute to a molecular switch important for survival that helps to guide the transition from immature to mature synaptic connections, as well as synapse regrowth and remodeling after neural injury.
Figures



References
-
- Bark IC (1993) Structure of the chicken gene for SNAP-25 reveals duplicated exon encoding distinct isoforms of the protein. J Mol Biol 233: 67-76. - PubMed
-
- Bark IC, Wilson MC (1994) Human cDNA clones encoding two different isoforms of the nerve terminal protein SNAP-25. Gene 139: 291-292. - PubMed
-
- Bock JB, Matern HT, Peden AA, Scheller RH (2001) A genomic perspective on membrane compartment organization. Nature 409: 839-841. - PubMed
-
- Boschert U, O'Shaughnessy C, Dickinson R, Tessari M, Bendotti C, Catsicas S, Pich EM (1996) Developmental and plasticity-related differential expression of two SNAP-25 isoforms in the rat brain. J Comp Neurol 367: 177-193. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
