Transcriptional response of Candida albicans upon internalization by macrophages
- PMID: 15470236
- PMCID: PMC522606
- DOI: 10.1128/EC.3.5.1076-1087.2004
Transcriptional response of Candida albicans upon internalization by macrophages
Abstract
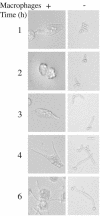
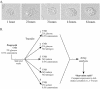
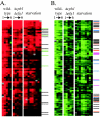
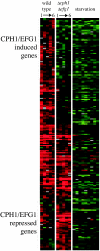
The opportunistic fungal pathogen Candida albicans is both a benign gut commensal and a frequently fatal systemic pathogen. The interaction of C. albicans with the host's innate immune system is the primary factor in this balance; defects in innate immunity predispose the patient to disseminated candidiasis. Because of the central importance of phagocytic cells in defense against fungal infections, we have investigated the response of C. albicans to phagocytosis by mammalian macrophages using genomic transcript profiling. This analysis reveals a dramatic reprogramming of transcription in C. albicans that occurs in two successive steps. In the early phase cells shift to a starvation mode, including gluconeogenic growth, activation of fatty acid degradation, and downregulation of translation. In a later phase, as hyphal growth enables C. albicans to escape from the macrophage, cells quickly resume glycolytic growth. In addition, there is a substantial nonmetabolic response imbedded in the early phase, including machinery for DNA damage repair, oxidative stress responses, peptide uptake systems, and arginine biosynthesis. Further, a surprising percentage of the genes that respond specifically to macrophage contact have no known homologs, suggesting that the organism has undergone substantial evolutionary adaptations to the commensal or pathogen lifestyle. This transcriptional reprogramming is almost wholly absent in the related, but nonpathogenic, yeast Saccharomyces cerevisiae, suggesting that these large-scale and coordinated changes contribute significantly to the ability of this organism to survive and cause disease in vivo.
Figures



References
-
- Ausubel, F. M., B. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2000. Current protocols in molecular biology. John Wiley & Sons, Edison, N.J.
-
- Braun, B. R., and A. D. Johnson. 1997. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277:105-109. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

