Macrodomain organization of the Escherichia coli chromosome
- PMID: 15470498
- PMCID: PMC524398
- DOI: 10.1038/sj.emboj.7600434
Macrodomain organization of the Escherichia coli chromosome
Abstract
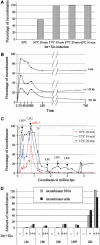
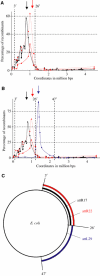
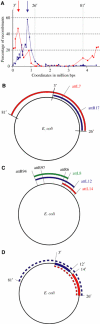
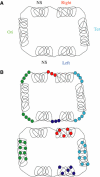
We have explored the Escherichia coli chromosome architecture by genetic dissection, using a site-specific recombination system that reveals the spatial proximity of distant DNA sites and records interactions. By analysing the percentages of recombination between pairs of sites scattered over the chromosome, we observed that DNA interactions were restricted to within subregions of the chromosome. The results indicated an organization into a ring composed of four macrodomains and two less-structured regions. Two of the macrodomains defined by recombination efficiency are similar to the Ter and Ori macrodomains observed by FISH. Two newly characterized macrodomains flank the Ter macrodomain and two less-structured regions flank the Ori macrodomain. Also the interactions between sister chromatids are rare, suggesting that chromosome segregation quickly follows replication. These results reveal structural features that may be important for chromosome dynamics during the cell cycle.
Figures


References
-
- Azaro MA, Landy A (2002) Integrase and the Int family. In Mobile DNA II, Craig NL, Craigie R, Gellert M, Lambowitz AM (eds) pp 118–148. Washington, DC: ASM Press
-
- Ben-Yehuda S, Rudner DZ, Losick R (2003) RacA, a bacterial protein that anchors chromosomes to the cell poles. Science 299: 532–536 - PubMed
-
- Capiaux H, Cornet F, Corre J, Guijo MI, Perals K, Rebollo JE, Louarn JM (2001) Polarization of the Escherichia coli chromosome. A view from the terminus. Biochimie 83: 161–170 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

