Inefficient degradation of truncated polyglutamine proteins by the proteasome
- PMID: 15470501
- PMCID: PMC524390
- DOI: 10.1038/sj.emboj.7600426
Inefficient degradation of truncated polyglutamine proteins by the proteasome
Abstract
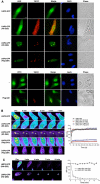
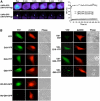
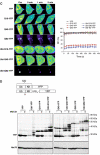
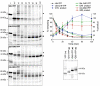
Accumulation of mutant proteins into misfolded species and aggregates is characteristic for diverse neurodegenerative diseases including the polyglutamine diseases. While several studies have suggested that polyglutamine protein aggregates impair the ubiquitin-proteasome system, the molecular mechanisms underlying the interaction between polyglutamine proteins and the proteasome have remained elusive. In this study, we use fluorescence live-cell imaging to demonstrate that the proteasome is sequestered irreversibly within aggregates of overexpressed N-terminal mutant Huntingtin fragment or simple polyglutamine expansion proteins. Moreover, by direct targeting of polyglutamine proteins for proteasomal degradation, we observe incomplete degradation of these substrates both in vitro and in vivo. Thus, our data reveal that intrinsic properties of the polyglutamine proteins prevent their efficient degradation and clearance. Additionally, fluorescence resonance energy transfer is detected between the proteasome and aggregated polyglutamine proteins indicative of a close and stable interaction. We propose that polyglutamine-containing proteins are kinetically trapped within proteasomes, which could explain their deleterious effects on cellular function over time.
Figures
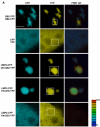
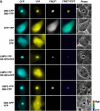
References
-
- Bence NF, Sampat RM, Kopito RR (2001) Impairment of the ubiquitin–proteasome system by protein aggregation. Science 292: 1552–1555 - PubMed
-
- Chen HK, Fernandez-Funez P, Acevedo SF, Lam YC, Kaytor MD, Fernandez MH, Aitken A, Skoulakis EM, Orr HT, Botas J, Zoghbi HY (2003) Interaction of Akt-phosphorylated ataxin-1 with 14-3-3 mediates neurodegeneration in spinocerebellar ataxia type 1. Cell 113: 457–468 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

