Colocalization of L-phenylalanine ammonia-lyase and cinnamate 4-hydroxylase for metabolic channeling in phenylpropanoid biosynthesis
- PMID: 15472080
- PMCID: PMC527201
- DOI: 10.1105/tpc.104.024406
Colocalization of L-phenylalanine ammonia-lyase and cinnamate 4-hydroxylase for metabolic channeling in phenylpropanoid biosynthesis
Abstract
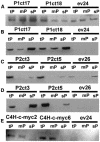
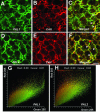
Metabolic channeling has been proposed to occur at the entry point into plant phenylpropanoid biosynthesis. To determine whether isoforms of L-Phe ammonia-lyase (PAL), the first enzyme in the pathway, can associate with the next enzyme, the endomembrane-bound cinnamate 4-hydroxylase (C4H), to facilitate channeling, we generated transgenic tobacco (Nicotiana tabacum) plants independently expressing epitope-tagged versions of two PAL isoforms (PAL1 and PAL2) and C4H. Subcellular fractionation and protein gel blot analysis using epitope- and PAL isoform-specific antibodies indicated both microsomal and cytosolic locations of PAL1 but only cytosolic localization of PAL2. However, both PAL isoforms were microsomally localized in plants overexpressing C4H. These results, which suggest that C4H itself may organize the complex for membrane association of PAL, were confirmed using PAL-green fluorescent protein (GFP) fusions with localization by confocal microscopy. Coexpression of unlabeled PAL1 with PAL2-GFP resulted in a shift of fluorescence localization from endomembranes to cytosol in C4H overexpressing plants, whereas coexpression of unlabeled PAL2 with PAL1-GFP did not affect PAL1-GFP localization, indicating that PAL1 has a higher affinity for its membrane localization site than does PAL2. Dual-labeling immunofluorescence and fluorescence energy resonance transfer (FRET) studies confirmed colocalization of PAL and C4H. However, FRET analysis with acceptor photobleaching suggested that the colocalization was not tight.
Figures





References
-
- Allwood, E.G., Davies, D.R., Gerrish, C., Ellis, B.E., and Bolwell, G.P. (1999). Phosphorylation of phenylalanine ammonia-lyase: Evidence for a novel protein kinase and identification of the phosphorylated residue. FEBS Lett. 457, 47–52. - PubMed
-
- Bak, S., Kahn, R.A., Nielsen, H.L., Moller, B.L., and Halkier, B.A. (1998). Cloning of three A-type cytochromes P450, CYP71E1, CYP98, and CYP99 from Sorghum bicolor (L.) Moench by a PCR approach and identification by expression in Escherichia coli of CYP71E1 as a multifunctional cytochrome P450 in the biosynthesis of the cyanogenic glucoside dhurrin. Plant Mol. Biol. 36, 393–405. - PubMed
-
- Bolwell, G.P. (1992). A role for phosphorylation in the down-regulation of phenylalanine ammonia-lyase in suspension-cultured cells of french bean. Phytochemistry 31, 4081–4086.
-
- Bolwell, G.P., Sap, J., Cramer, C.L., Schuch, W., Lamb, C.J., and Dixon, R.A. (1985). L-Phenylalanine ammonia-lyase from Phaseolus vulgaris: Partial degradation of enzyme subunits in vitro and in vivo. Biochim. Biophys. Acta 881, 210–221.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

