Comparative analysis of Gtf isozyme production and diversity in isolates of Streptococcus mutans with different biofilm growth phenotypes
- PMID: 15472313
- PMCID: PMC522304
- DOI: 10.1128/JCM.42.10.4586-4592.2004
Comparative analysis of Gtf isozyme production and diversity in isolates of Streptococcus mutans with different biofilm growth phenotypes
Abstract
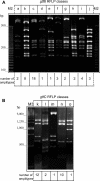
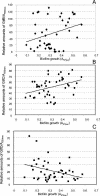
Streptococcus mutans is the main pathogenic agent of dental caries. Glucosyltransferases (Gtfs) produced by these bacteria are important virulence factors because they catalyze the extracellular synthesis of glucans that are necessary for bacterial accumulation in the dental biofilm. The diversity of GtfB and GtfC isozymes was analyzed in 44 genotypes of S. mutans that showed a range of abilities to form biofilms in vitro. Several approaches were used to characterize these isozymes, including restriction fragment length polymorphism analysis of the gtfB and gtfC genes, zymographic analysis of the identified GtfB and GtfC genotypes, and quantitation of isozyme production in immunoblot experiments with specific monoclonal antibodies. A high diversity of gtf genes, patterns of enzymatic activity, and isozyme production was identified among the isolates tested. GtfC and, to a lesser extent, GtfB were produced in significantly higher amounts by strains that had high biofilm-forming ability than by strains with low biofilm-forming ability. Biofilm formation was independent of the GtfB and GtfC genotype. Atypical strains that showed an apparent single Gtf isozyme of intermediate size between GtfB and GtfC were also identified. The results indicate that various expression levels of GtfB and GtfC isozymes are associated with the ability of distinct S. mutans genotypes to grow as biofilms, strengthening the results of previous genetic and biochemical studies performed with laboratory strains. These studies also emphasize the need to identify factors that control gtf gene expression.
Figures


Similar articles
-
Differential expression profiles of Streptococcus mutans ftf, gtf and vicR genes in the presence of dietary carbohydrates at early and late exponential growth phases.Carbohydr Res. 2006 Sep 4;341(12):2090-7. doi: 10.1016/j.carres.2006.05.010. Epub 2006 Jun 9. Carbohydr Res. 2006. PMID: 16764842
-
Transcriptional analysis of gtfB, gtfC, and gbpB and their putative response regulators in several isolates of Streptococcus mutans.Oral Microbiol Immunol. 2008 Dec;23(6):466-73. doi: 10.1111/j.1399-302X.2008.00451.x. Oral Microbiol Immunol. 2008. PMID: 18954352
-
Inhibitory effect of Lactobacillus salivarius on Streptococcus mutans biofilm formation.Mol Oral Microbiol. 2015 Feb;30(1):16-26. doi: 10.1111/omi.12063. Epub 2014 Sep 8. Mol Oral Microbiol. 2015. PMID: 24961744
-
Biology of Streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms.Caries Res. 2011;45(1):69-86. doi: 10.1159/000324598. Epub 2011 Feb 23. Caries Res. 2011. PMID: 21346355 Free PMC article. Review.
-
Molecular mechanisms of inhibiting glucosyltransferases for biofilm formation in Streptococcus mutans.Int J Oral Sci. 2021 Sep 30;13(1):30. doi: 10.1038/s41368-021-00137-1. Int J Oral Sci. 2021. PMID: 34588414 Free PMC article. Review.
Cited by
-
Downregulation of GbpB, a component of the VicRK regulon, affects biofilm formation and cell surface characteristics of Streptococcus mutans.Infect Immun. 2011 Feb;79(2):786-96. doi: 10.1128/IAI.00725-10. Epub 2010 Nov 15. Infect Immun. 2011. PMID: 21078847 Free PMC article.
-
Exploring the Genomic Diversity and Cariogenic Differences of Streptococcus mutans Strains Through Pan-Genome and Comparative Genome Analysis.Curr Microbiol. 2017 Oct;74(10):1200-1209. doi: 10.1007/s00284-017-1305-z. Epub 2017 Jul 17. Curr Microbiol. 2017. PMID: 28717847
-
Mechanistic, genomic and proteomic study on the effects of BisGMA-derived biodegradation product on cariogenic bacteria.Dent Mater. 2017 Feb;33(2):175-190. doi: 10.1016/j.dental.2016.11.007. Epub 2016 Dec 3. Dent Mater. 2017. PMID: 27919444 Free PMC article.
-
In vitro manganese-dependent cross-talk between Streptococcus mutans VicK and GcrR: implications for overlapping stress response pathways.PLoS One. 2014 Dec 23;9(12):e115975. doi: 10.1371/journal.pone.0115975. eCollection 2014. PLoS One. 2014. PMID: 25536343 Free PMC article.
-
Whole genome sequence and phenotypic characterization of a Cbm+ serotype e strain of Streptococcus mutans.Mol Oral Microbiol. 2018 Jun;33(3):257-269. doi: 10.1111/omi.12222. Epub 2018 Apr 17. Mol Oral Microbiol. 2018. PMID: 29524318 Free PMC article.
References
-
- Childers, N. K., S. S. Zhang, and S. M. Michalek. 1994. Oral immunization of humans with dehydrated liposomes containing Streptococcus mutans glucosyltransferase induces salivary immunoglobulin A2 antibody responses. Oral Microbiol. Immunol. 9:146-153. - PubMed
-
- Fujiwara, T., Y. Terao, T. Hoshino, S. Kawabata, T. Ooshima, S. Sobue, S. Kimura, and S. Hamada. 1998. Molecular analyses of glucosyltransferase genes among strains of Streptococcus mutans. FEMS Microbiol. Lett. 161:331-336. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

