Content-rich biological network constructed by mining PubMed abstracts
- PMID: 15473905
- PMCID: PMC528731
- DOI: 10.1186/1471-2105-5-147
Content-rich biological network constructed by mining PubMed abstracts
Abstract
Background: The integration of the rapidly expanding corpus of information about the genome, transcriptome, and proteome, engendered by powerful technological advances, such as microarrays, and the availability of genomic sequence from multiple species, challenges the grasp and comprehension of the scientific community. Despite the existence of text-mining methods that identify biological relationships based on the textual co-occurrence of gene/protein terms or similarities in abstract texts, knowledge of the underlying molecular connections on a large scale, which is prerequisite to understanding novel biological processes, lags far behind the accumulation of data. While computationally efficient, the co-occurrence-based approaches fail to characterize (e.g., inhibition or stimulation, directionality) biological interactions. Programs with natural language processing (NLP) capability have been created to address these limitations, however, they are in general not readily accessible to the public.
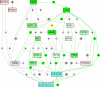
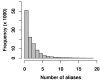
Results: We present a NLP-based text-mining approach, Chilibot, which constructs content-rich relationship networks among biological concepts, genes, proteins, or drugs. Amongst its features, suggestions for new hypotheses can be generated. Lastly, we provide evidence that the connectivity of molecular networks extracted from the biological literature follows the power-law distribution, indicating scale-free topologies consistent with the results of previous experimental analyses.
Conclusions: Chilibot distills scientific relationships from knowledge available throughout a wide range of biological domains and presents these in a content-rich graphical format, thus integrating general biomedical knowledge with the specialized knowledge and interests of the user. Chilibot http://www.chilibot.net can be accessed free of charge to academic users.
Figures


References
-
- Stapley BJ, Benoit G. Biobibliometrics: information retrieval and visualization from co-occurrences of gene names in Medline abstracts. Pac Symp Biocomput. 2000:529–540. - PubMed
-
- Donaldson I, Martin J, de Bruijn B, Wolting C, Lay V, Tuekam B, Zhang S, Baskin B, Bader GD, Michalickova K. PreBIND and Textomy – mining the biomedical literature for protein-protein interactions using a support vector machine. BMC Bioinformatics. 2003;4:11. doi: 10.1186/1471-2105-4-11. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

