KpnBI is the prototype of a new family (IE) of bacterial type I restriction-modification system
- PMID: 15475385
- PMCID: PMC524312
- DOI: 10.1093/nar/gnh134
KpnBI is the prototype of a new family (IE) of bacterial type I restriction-modification system
Abstract
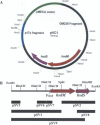
KpnBI is a restriction-modification (R-M) system recognized in the GM236 strain of Klebsiella pneumoniae. Here, the KpnBI modification genes were cloned into a plasmid using a modification expression screening method. The modification genes that consist of both hsdM (2631 bp) and hsdS (1344 bp) genes were identified on an 8.2 kb EcoRI chromosomal fragment. These two genes overlap by one base and share the same promoter located upstream of the hsdM gene. Using recently developed plasmid R-M tests and a computer program RM Search, the DNA recognition sequence for the KpnBI enzymes was identified as a new 8 nt sequence containing one degenerate base with a 6 nt spacer, CAAANNNNNNRTCA. From Dam methylation and HindIII sensitivity tests, the methylation loci were predicted to be the italicized third adenine in the 5' specific region and the adenine opposite the italicized thymine in the 3' specific region. Combined with previous sequence data for hsdR, we concluded that the KpnBI system is a typical type I R-M system. The deduced amino acid sequences of the three subunits of the KpnBI system show only limited homologies (25 to 33% identity) at best, to the four previously categorized type I families (IA, IB, IC, and ID). Furthermore, their identity scores to other uncharacterized putative genome type I sequences were 53% at maximum. Therefore, we propose that KpnBI is the prototype of a new 'type IE' family.
Figures




References
-
- Xu G., Willert,J., Kapfer,W. and Trautner,T.A. (1995) BsuCI, a type-I restriction-modification system in Bacillus subtilis. Gene, 157, 59. - PubMed
-
- Hadi S.M., Bachi,B., Shepherd,J.C., Yuan,R., Ineichen,K. and Bickle,T.A. (1979) DNA recognition and cleavage by the EcoP15 restriction endonuclease. J. Mol. Biol., 134, 655–666. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

