Evolutionary relationships of Aurora kinases: implications for model organism studies and the development of anti-cancer drugs
- PMID: 15476560
- PMCID: PMC524484
- DOI: 10.1186/1471-2148-4-39
Evolutionary relationships of Aurora kinases: implications for model organism studies and the development of anti-cancer drugs
Abstract
Background: As key regulators of mitotic chromosome segregation, the Aurora family of serine/threonine kinases play an important role in cell division. Abnormalities in Aurora kinases have been strongly linked with cancer, which has lead to the recent development of new classes of anti-cancer drugs that specifically target the ATP-binding domain of these kinases. From an evolutionary perspective, the species distribution of the Aurora kinase family is complex. Mammals uniquely have three Aurora kinases, Aurora-A, Aurora-B, and Aurora-C, while for other metazoans, including the frog, fruitfly and nematode, only Aurora-A and Aurora-B kinases are known. The fungi have a single Aurora-like homolog. Based on the tacit assumption of orthology to human counterparts, model organism studies have been central to the functional characterization of Aurora kinases. However, the ortholog and paralog relationships of these kinases across various species have not been rigorously examined. Here, we present comprehensive evolutionary analyses of the Aurora kinase family.
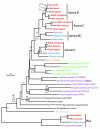
Results: Phylogenetic trees suggest that all three vertebrate Auroras evolved from a single urochordate ancestor. Specifically, Aurora-A is an orthologous lineage in cold-blooded vertebrates and mammals, while structurally similar Aurora-B and Aurora-C evolved more recently in mammals from a duplication of an ancestral Aurora-B/C gene found in cold-blooded vertebrates. All so-called Aurora-A and Aurora-B kinases of non-chordates are ancestral to the clade of chordate Auroras and, therefore, are not strictly orthologous to vertebrate counterparts. Comparisons of human Aurora-B and Aurora-C sequences to the resolved 3D structure of human Aurora-A lends further support to the evolutionary scenario that vertebrate Aurora-B and Aurora-C are closely related paralogs. Of the 26 residues lining the ATP-binding active site, only three were variant and all were specific to Aurora-A.
Conclusions: In this study, we found that invertebrate Aurora-A and Aurora-B kinases are highly divergent protein families from their chordate counterparts. Furthermore, while the Aurora-A family is ubiquitous among all vertebrates, the Aurora-B and Aurora-C families in humans arose from a gene duplication event in mammals. These findings show the importance of understanding evolutionary relationships in the interpretation and transference of knowledge from studies of model organism systems to human cellular biology. In addition, given the important role of Aurora kinases in cancer, evolutionary analysis and comparisons of ATP-binding domains suggest a rationale for designing dual action anti-tumor drugs that inhibit both Aurora-B and Aurora-C kinases.
Figures


References
-
- Schumacher JM, Golden A, Donovan PJ. AIR-2: An Aurora/Ipl1-related protein kinase associated with chromosomes and midbody microtubules is required for polar body extrusion and cytokinesis in Caenorhabditis elegans embryos. J Cell Biol. 1998;143:1635–1646. doi: 10.1083/jcb.143.6.1635. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

