Microfluidic PicoArray synthesis of oligodeoxynucleotides and simultaneous assembling of multiple DNA sequences
- PMID: 15477391
- PMCID: PMC524290
- DOI: 10.1093/nar/gkh879
Microfluidic PicoArray synthesis of oligodeoxynucleotides and simultaneous assembling of multiple DNA sequences
Abstract
Large DNA constructs of arbitrary sequences can currently be assembled with relative ease by joining short synthetic oligodeoxynucleotides (oligonucleotides). The ability to mass produce these synthetic genes readily will have a significant impact on research in biology and medicine. Presently, high-throughput gene synthesis is unlikely, due to the limits of oligonucleotide synthesis. We describe a microfluidic PicoArray method for the simultaneous synthesis and purification of oligonucleotides that are designed for multiplex gene synthesis. Given the demand for highly pure oligonucleotides in gene synthesis processes, we used a model to improve key reaction steps in DNA synthesis. The oligonucleotides obtained were successfully used in ligation under thermal cycling conditions to generate DNA constructs of several hundreds of base pairs. Protein expression using the gene thus synthesized was demonstrated. We used a DNA assembly strategy, i.e. ligation followed by fusion PCR, and achieved effective assembling of up to 10 kb DNA constructs. These results illustrate the potential of microfluidics-based ultra-fast oligonucleotide parallel synthesis as an enabling tool for modern synthetic biology applications, such as the construction of genome-scale molecular clones and cell-free large scale protein expression.
Figures



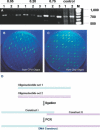

References
-
- Stemmer W.P.C., Crameri,A., Ha,K.D., Brennan,T.M. and Heyneker,H.L. (1995) Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene, 164, 49–53. - PubMed
-
- Dillon P.J. and Rosen,C.A. (1990) A rapid method for the construction of synthetic genes using the polymerase chain reaction. Biotechniques, 9, 298–300. - PubMed
-
- Bambot S.B. and Russell,A.J. (1993) Efficient total gene synthesis of 1.35-kb hybrid alpha-lytic protease gene using the polymerase chain reaction. PCR Methods Appl., 2, 266–271. - PubMed
-
- Sandhu G.S. Aleff,R.A. and Kline,B.C. (1992) Dual asymmetric PCR: one-step construction of synthetic genes. Biotechniques, 12, 14–16. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

