Intracellular localization and protein interactions of the gene 1 protein p28 during mouse hepatitis virus replication
- PMID: 15479796
- PMCID: PMC523235
- DOI: 10.1128/JVI.78.21.11551-11562.2004
Intracellular localization and protein interactions of the gene 1 protein p28 during mouse hepatitis virus replication
Abstract
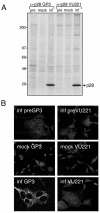
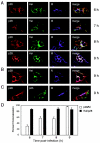
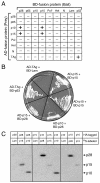
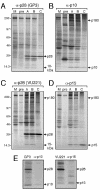
Coronaviruses encode the largest replicase polyprotein of any known positive-strand RNA virus. Replicase protein precursors and mature products are thought to mediate the formation and function of viral replication complexes on the surfaces of intracellular double-membrane vesicles. However, the functions of only a few of these proteins are known. For the coronavirus mouse hepatitis virus (MHV), the first proteolytic processing event of the replicase polyprotein liberates an amino-terminal 28-kDa product (p28). While previous biochemical studies have suggested that p28 is associated with viral replication complexes, the intracellular localization and interactions of p28 with other proteins during the course of MHV replication have not been defined. We used immunofluorescence confocal microscopy to show that p28 localizes to viral replication complexes in the cytoplasm during early times postinfection. However, at late times postinfection, p28 localizes to sites of M accumulation distinct from the replication complex. Furthermore, by yeast two-hybrid and coimmunoprecipitation analyses, we demonstrate that p28 specifically binds to p10 and p15, two coronavirus replicase proteins of unknown function. Deletion mutagenesis experiments determined that the carboxy terminus of p28 is not required for its interactions with p10 and p15. These results suggest that p28 may play a part at the replication complex by interacting with p10 and p15. Moreover, our findings highlight a potential role for p28 at virion assembly sites.
Figures



References
-
- Bi, W., J. D. Pinon, S. Hughes, P. J. Bonilla, K. V. Holmes, S. R. Weiss, and J. L. Leibowitz. 1998. Localization of mouse hepatitis virus open reading frame 1A derived proteins. J. Neurovirol. 4:594-605. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

