Genome-wide analyses of avian sarcoma virus integration sites
- PMID: 15479807
- PMCID: PMC523270
- DOI: 10.1128/JVI.78.21.11656-11663.2004
Genome-wide analyses of avian sarcoma virus integration sites
Abstract
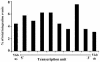
The chromosomal features that influence retroviral integration site selection are not well understood. Here, we report the mapping of 226 avian sarcoma virus (ASV) integration sites in the human genome. The results show that the sites are distributed over all chromosomes, and no global bias for integration site selection was detected. However, RNA polymerase II transcription units (protein-encoding genes) appear to be favored targets of ASV integration. The integration frequency within genes is similar to that previously described for murine leukemia virus but distinct from the higher frequency observed with human immunodeficiency virus type 1. We found no evidence for preferred ASV integration sites over the length of genes and immediate flanking regions. Microarray analysis of uninfected HeLa cells revealed that the expression levels of ASV target genes were similar to the median level for all genes represented in the array. Although expressed genes were targets for integration, we found no preference for integration into highly expressed genes. Our results provide a more detailed description of the chromosomal features that may influence ASV integration and support the idea that distinct, virus-specific mechanisms mediate integration site selection. Such differences may be relevant to viral pathogenesis and provide utility in retroviral vector design.
Figures
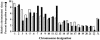


References
-
- Brown, P. O., B. Bowerman, H. E. Varmus, and J. M. Bishop. 1987. Correct integration of retroviral DNA in vitro. Cell 49:347-356. - PubMed
-
- Bukrinsky, M. I., N. Sharova, T. L. McDonald, T. Pushkarskaya, W. G. Tarpley, and M. Stevenson. 1993. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc. Natl. Acad. Sci. USA 90:6125-6129. - PMC - PubMed
-
- Bushman, F. D. 1999. Host proteins in retroviral cDNA integration. Adv. Virus Res. 52:301-317. - PubMed
-
- Bushman, F. D. 2003. Targeting survival: integration site selection by retroviruses and LTR-retrotransposons. Cell 115:135-138. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

