Structural and thermodynamic basis for the binding of TMC114, a next-generation human immunodeficiency virus type 1 protease inhibitor
- PMID: 15479840
- PMCID: PMC523255
- DOI: 10.1128/JVI.78.21.12012-12021.2004
Structural and thermodynamic basis for the binding of TMC114, a next-generation human immunodeficiency virus type 1 protease inhibitor
Abstract
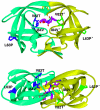
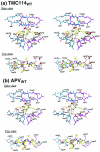
TMC114, a newly designed human immunodeficiency virus type 1 (HIV-1) protease inhibitor, is extremely potent against both wild-type (wt) and multidrug-resistant (MDR) viruses in vitro as well as in vivo. Although chemically similar to amprenavir (APV), the potency of TMC114 is substantially greater. To examine the basis for this potency, we solved crystal structures of TMC114 complexed with wt HIV-1 protease and TMC114 and APV complexed with an MDR (L63P, V82T, and I84V) protease variant. In addition, we determined the corresponding binding thermodynamics by isothermal titration calorimetry. TMC114 binds approximately 2 orders of magnitude more tightly to the wt enzyme (K(d) = 4.5 x 10(-12) M) than APV (K(d) = 3.9 x 10(-10) M). Our X-ray data (resolution ranging from 2.2 to 1.2 A) reveal strong interactions between the bis-tetrahydrofuranyl urethane moiety of TMC114 and main-chain atoms of D29 and D30. These interactions appear largely responsible for TMC114's very favorable binding enthalpy to the wt protease (-12.1 kcal/mol). However, TMC114 binding to the MDR HIV-1 protease is reduced by a factor of 13.3, whereas the APV binding constant is reduced only by a factor of 5.1. However, even with the reduction in binding affinity to the MDR HIV protease, TMC114 still binds with an affinity that is more than 1.5 orders of magnitude tighter than the first-generation inhibitors. Both APV and TMC114 fit predominantly within the substrate envelope, a property that may be associated with decreased susceptibility to drug-resistant mutations relative to that of first-generation inhibitors. Overall, TMC114's potency against MDR viruses is likely a combination of its extremely high affinity and close fit within the substrate envelope.
Figures




References
-
- Ala, P. J., E. E. Huston, R. M. Klabe, P. K. Jadhav, P. Y. Lam, and C. H. Chang. 1998. Counteracting HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with XV638 and SD146, cyclic urea amides with broad specificities. Biochemistry 37:15042-15049. - PubMed
-
- Arasteh, K., A. Clumeck, H. Pozniak, H. Jäger, M. De Pauw, H. Müller, M. Peeters, R. Hoetelmans, S. De Meyer, I. Van der Sandt, S. Comhaire, and R. Van der Geest. 2003. First clinical results on antiretroviral activity, pharmacokinetics, and safety of TMC114, an HIV-1 protease inhibitor, in multiple PI-experienced patients, abstr. 8. 10th Conference on Retroviruses and Opportunistic Infections (CROI), Boston, Mass.
-
- Baldwin, E. T., T. N. Bhat, B. Liu, N. Pattabiraman, and J. W. Erickson. 1995. Structural basis of drug resistance for the V82A mutant of HIV-1 proteinase. Nat. Struct. Biol. 2:244-249. - PubMed
-
- Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905-921. - PubMed
-
- Chapman, M. S. 1995. Restrained real-space macromolecular refinement using a new resolution-dependent electron density function. Acta Crystallogr. A 51:69-80.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

