Genetic interactions between hepatitis C virus replicons
- PMID: 15479852
- PMCID: PMC523274
- DOI: 10.1128/JVI.78.21.12085-12089.2004
Genetic interactions between hepatitis C virus replicons
Abstract
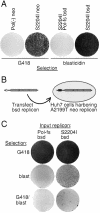
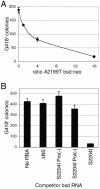
To investigate interactions between hepatitis C virus (HCV) RNA replication complexes, a system was developed to simultaneously select different HCV subgenomic replicons within the same cell. Transcomplementation of defective replicons was not observed, suggesting an isolated and independent nature of the HCV RNA replication complex. In contrast, a high level of competition between replicons was observed, such that the presence and increased fitness of one replicon reduced the capacity of a second one to stably replicate. These results suggest that at least one factor in Huh7 cells required for HCV RNA replication is limiting and saturable.
Figures


Similar articles
-
Persistent replication of hepatitis C virus replicons expressing the beta-lactamase reporter in subpopulations of highly permissive Huh7 cells.J Virol. 2003 Mar;77(5):2928-35. doi: 10.1128/jvi.77.5.2928-2935.2003. J Virol. 2003. PMID: 12584317 Free PMC article.
-
Hepatitis C virus subgenomic replicons in the human embryonic kidney 293 cell line.J Virol. 2004 Jan;78(1):491-501. doi: 10.1128/jvi.78.1.491-501.2004. J Virol. 2004. PMID: 14671129 Free PMC article.
-
Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells.J Virol. 2002 Mar;76(6):2997-3006. doi: 10.1128/jvi.76.6.2997-3006.2002. J Virol. 2002. PMID: 11861865 Free PMC article.
-
Replication studies using genotype 1a subgenomic hepatitis C virus replicons.J Virol. 2003 May;77(9):5352-9. doi: 10.1128/jvi.77.9.5352-5359.2003. J Virol. 2003. PMID: 12692237 Free PMC article.
-
Dominant negative effect of wild-type NS5A on NS5A-adapted subgenomic hepatitis C virus RNA replicon.J Gen Virol. 2004 Jul;85(Pt 7):1867-1875. doi: 10.1099/vir.0.80006-0. J Gen Virol. 2004. PMID: 15218171
Cited by
-
Distinct functions of NS5A in hepatitis C virus RNA replication uncovered by studies with the NS5A inhibitor BMS-790052.J Virol. 2011 Jul;85(14):7312-20. doi: 10.1128/JVI.00253-11. Epub 2011 May 18. J Virol. 2011. PMID: 21593143 Free PMC article.
-
Hepatitis C virus subgenomic replicon requires an active NS3 RNA helicase.J Virol. 2006 Jan;80(1):404-11. doi: 10.1128/JVI.80.1.404-411.2006. J Virol. 2006. PMID: 16352565 Free PMC article.
-
Characterization of determinants important for hepatitis C virus p7 function in morphogenesis by using trans-complementation.J Virol. 2009 Nov;83(22):11682-93. doi: 10.1128/JVI.00691-09. Epub 2009 Sep 2. J Virol. 2009. PMID: 19726506 Free PMC article.
-
Studying hepatitis C virus: making the best of a bad virus.J Virol. 2007 Sep;81(17):8853-67. doi: 10.1128/JVI.00753-07. Epub 2007 May 23. J Virol. 2007. PMID: 17522203 Free PMC article. Review. No abstract available.
-
Autophagy in hepatitis C virus-host interactions: potential roles and therapeutic targets for liver-associated diseases.World J Gastroenterol. 2014 May 21;20(19):5773-93. doi: 10.3748/wjg.v20.i19.5773. World J Gastroenterol. 2014. PMID: 24914338 Free PMC article. Review.
References
-
- Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. - PubMed
-
- Colina, R., D. Casane, S. Vasquez, L. Garcia-Aguirre, A. Chunga, H. Romero, B. Khan, and J. Cristina. 2004. Evidence of intratypic recombination in natural populations of hepatitis C virus. J. Gen. Virol. 85:31-37. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

