Gene prediction and verification in a compact genome with numerous small introns
- PMID: 15479946
- PMCID: PMC525692
- DOI: 10.1101/gr.2816704
Gene prediction and verification in a compact genome with numerous small introns
Abstract
The genomes of clusters of related eukaryotes are now being sequenced at an increasing rate, creating a need for accurate, low-cost annotation of exon-intron structures. In this paper, we demonstrate that reverse transcription-polymerase chain reaction (RT-PCR) and direct sequencing based on predicted gene structures satisfy this need, at least for single-celled eukaryotes. The TWINSCAN gene prediction algorithm was adapted for the fungal pathogen Cryptococcus neoformans by using a precise model of intron lengths in combination with ungapped alignments between the genome sequences of the two closely related Cryptococcus varieties. This approach resulted in approximately 60% of known genes being predicted exactly right at every coding base and splice site. When previously unannotated TWINSCAN predictions were tested by RT-PCR and direct sequencing, 75% of targets spanning two predicted introns were amplified and produced high-quality sequence. When targets spanning the complete predicted open reading frame were tested, 72% of them amplified and produced high-quality sequence. We conclude that sequencing a small number of expressed sequence tags (ESTs) to provide training data, running TWINSCAN on an entire genome, and then performing RT-PCR and direct sequencing on all of its predictions would be a cost-effective method for obtaining an experimentally verified genome annotation.
Figures

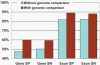
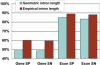

References
WEB SITE REFERENCES
-
- http://www.ncbi.nlm.nih.gov/Traces/; NCBI Trace Archive.
-
- http://genes.cse.wustl.edu/tenney-04-crypto-data/; Supplemental data for this paper.
-
- http://genes.cse.wustl.edu/; TWINSCAN home page, application, source code, and gene predictions.
-
- http://micro-gen.ouhsc.edu; Oklahoma University Health Sciences Center.
Publication types
MeSH terms
Grants and funding
- F33 HG002635/HG/NHGRI NIH HHS/United States
- R01-AI051209/AI/NIAID NIH HHS/United States
- R01 GM066303/GM/NIGMS NIH HHS/United States
- K22 HG000045/HG/NHGRI NIH HHS/United States
- R01-GM66303/GM/NIGMS NIH HHS/United States
- R01 AI051209/AI/NIAID NIH HHS/United States
- T32 HG00045/HG/NHGRI NIH HHS/United States
- T32 HG000045/HG/NHGRI NIH HHS/United States
- R01 AI050184/AI/NIAID NIH HHS/United States
- F33 HG002653/HG/NHGRI NIH HHS/United States
- R01-AI50184/AI/NIAID NIH HHS/United States
- R01-AI49173/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
