Interactions of eukaryotic translation initiation factor 3 (eIF3) subunit NIP1/c with eIF1 and eIF5 promote preinitiation complex assembly and regulate start codon selection
- PMID: 15485912
- PMCID: PMC522265
- DOI: 10.1128/MCB.24.21.9437-9455.2004
Interactions of eukaryotic translation initiation factor 3 (eIF3) subunit NIP1/c with eIF1 and eIF5 promote preinitiation complex assembly and regulate start codon selection
Abstract
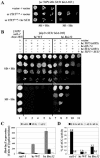
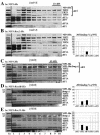
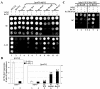
The N-terminal domain (NTD) of NIP1/eIF3c interacts directly with eIF1 and eIF5 and indirectly through eIF5 with the eIF2-GTP-Met-tRNA(i)(Met) ternary complex (TC) to form the multifactor complex (MFC). We investigated the physiological importance of these interactions by mutating 16 segments spanning the NIP1-NTD. Mutations in multiple segments reduced the binding of eIF1 or eIF5 to the NIP1-NTD. Mutating a C-terminal segment of the NIP1-NTD increased utilization of UUG start codons (Sui(-) phenotype) and was lethal in cells expressing eIF5-G31R that is hyperactive in stimulating GTP hydrolysis by the TC at AUG codons. Both effects of this NIP1 mutation were suppressed by eIF1 overexpression, as was the Sui(-) phenotype conferred by eIF5-G31R. Mutations in two N-terminal segments of the NIP1-NTD suppressed the Sui(-) phenotypes produced by the eIF1-D83G and eIF5-G31R mutations. From these and other findings, we propose that the NIP1-NTD coordinates an interaction between eIF1 and eIF5 that inhibits GTP hydrolysis at non-AUG codons. Two NIP1-NTD mutations were found to derepress GCN4 translation in a manner suppressed by overexpressing the TC, indicating that MFC formation stimulates TC recruitment to 40S ribosomes. Thus, the NIP1-NTD is required for efficient assembly of preinitiation complexes and also regulates the selection of AUG start codons in vivo.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
