The Mre11 nuclease is not required for 5' to 3' resection at multiple HO-induced double-strand breaks
- PMID: 15485933
- PMCID: PMC522228
- DOI: 10.1128/MCB.24.21.9682-9694.2004
The Mre11 nuclease is not required for 5' to 3' resection at multiple HO-induced double-strand breaks
Abstract
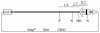
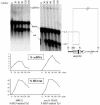
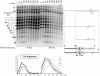
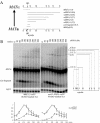
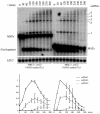
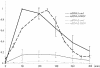
Current hypotheses suggest the Mre11 nuclease activity could be directly involved in double-strand break (DSB) resection in the presence of a large number of DSBs or limited to processing abnormal DNA ends. To distinguish between these possibilities, we used two methods to create large numbers of DSBs in Saccharomyces cerevisiae chromosomes, without introducing other substrates for the Mre11 nuclease. Multiple DSBs were created either by expressing the HO endonuclease in strains containing several HO cut sites embedded within randomly dispersed Ty1 elements or by phleomycin treatment. Analysis of resection by single-strand DNA formation in these systems showed no difference between strains containing MRE11 or the mre11-D56N nuclease defective allele, suggesting that the Mre11 nuclease is not involved in the extensive 5' to 3' resection of DSBs. We postulate that the ionizing radiation (IR) sensitivity of mre11 nuclease-defective mutants results from the accumulation of IR-induced DNA damage that is normally processed by the Mre11 nuclease. We also report that the processivity of 5' to 3' DSB resection and the yield of repaired products are affected by the number of DSBs in a dose-dependent manner. Finally, we show that the exonuclease Exo1 is involved in the processivity of 5' to 3' resection of an HO-induced DSB at the MAT locus.
Figures


References
-
- Alani, E., R. Padmore, and N. Kleckner. 1990. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell 61:419-436. - PubMed
-
- Amundsen, S. K., and G. R. Smith. 2003. Interchangeable parts of the Escherichia coli recombination machinery. Cell 112:741-744. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
