PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana
- PMID: 15486102
- PMCID: PMC527197
- DOI: 10.1105/tpc.104.025163
PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana
Abstract
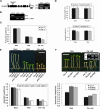
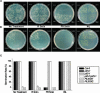
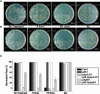
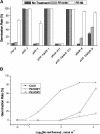
The first decision made by an angiosperm seed, whether to germinate or not, is based on integration of various environmental signals such as water and light. The phytochromes (Phys) act as red and far-red light (Pfr) photoreceptors to mediate light signaling through yet uncharacterized pathways. We report here that the PIF3-like 5 (PIL5) protein, a basic helix-loop-helix transcription factor, is a key negative regulator of phytochrome-mediated seed germination. PIL5 preferentially interacts with the Pfr forms of Phytochrome A (PhyA) and Phytochrome B (PhyB). Analyses of a pil5 mutant in conjunction with phyA and phyB mutants, a pif3 pil5 double mutant, and PIL5 overexpression lines indicate that PIL5 is a negative factor in Phy-mediated promotion of seed germination, inhibition of hypocotyl negative gravitropism, and inhibition of hypocotyl elongation. Our data identify PIL5 as the first Phy-interacting protein that regulates seed germination.
Figures


Similar articles
-
The Dof protein DAG1 mediates PIL5 activity on seed germination by negatively regulating GA biosynthetic gene AtGA3ox1.Plant J. 2010 Jan;61(2):312-23. doi: 10.1111/j.1365-313X.2009.04055.x. Epub 2009 Oct 26. Plant J. 2010. PMID: 19874540
-
A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors.Plant Cell. 2004 Nov;16(11):3033-44. doi: 10.1105/tpc.104.025643. Epub 2004 Oct 14. Plant Cell. 2004. PMID: 15486100 Free PMC article.
-
Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis.Plant J. 2006 Jul;47(1):124-39. doi: 10.1111/j.1365-313X.2006.02773.x. Epub 2006 Jun 1. Plant J. 2006. PMID: 16740147
-
Phytochrome-interacting factor from Arabidopsis to liverwort.Curr Opin Plant Biol. 2017 Feb;35:54-60. doi: 10.1016/j.pbi.2016.11.004. Epub 2016 Nov 19. Curr Opin Plant Biol. 2017. PMID: 27875778 Review.
-
Plant hormone signaling lightens up: integrators of light and hormones.Curr Opin Plant Biol. 2010 Oct;13(5):571-7. doi: 10.1016/j.pbi.2010.07.001. Epub 2010 Aug 23. Curr Opin Plant Biol. 2010. PMID: 20739215 Review.
Cited by
-
Integration of ABA, GA, and light signaling in seed germination through the regulation of ABI5.Front Plant Sci. 2022 Aug 24;13:1000803. doi: 10.3389/fpls.2022.1000803. eCollection 2022. Front Plant Sci. 2022. PMID: 36092418 Free PMC article. Review.
-
Phytochrome A in plants comprises two structurally and functionally distinct populations - water-soluble phyA' and amphiphilic phyA″.Biophys Rev. 2022 Jul 1;14(4):905-921. doi: 10.1007/s12551-022-00974-2. eCollection 2022 Aug. Biophys Rev. 2022. PMID: 36124260 Free PMC article. Review.
-
Genetic, Epigenetic, and Environmental Control of Seed Dormancy and Germination.Methods Mol Biol. 2024;2830:3-12. doi: 10.1007/978-1-0716-3965-8_1. Methods Mol Biol. 2024. PMID: 38977563
-
CROWN ROOTLESS1 binds DNA with a relaxed specificity and activates OsROP and OsbHLH044 genes involved in crown root formation in rice.Plant J. 2022 Jul;111(2):546-566. doi: 10.1111/tpj.15838. Epub 2022 Jul 9. Plant J. 2022. PMID: 35596715 Free PMC article.
-
The time of day effects of warm temperature on flowering time involve PIF4 and PIF5.J Exp Bot. 2014 Mar;65(4):1141-51. doi: 10.1093/jxb/ert487. J Exp Bot. 2014. PMID: 24574484 Free PMC article.
References
-
- Ahmad, M., and Cashmore, A.R. (1993). HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366, 162–166. - PubMed
-
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. - PubMed
-
- Alvarez, J., and Smyth, D.R. (1999). CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 126, 2377–2386. - PubMed
-
- Bauer, D., Viczian, A., Kircher, S., Nobis, T., Nitschke, R., Kunkel, T., Panigrahi, K.C., Adam, E., Fejes, E., Schafer, E., and Nagy, F. (2004). Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16, 1433–1445. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

