A gene regulatory network model for cell-fate determination during Arabidopsis thaliana flower development that is robust and recovers experimental gene expression profiles
- PMID: 15486106
- PMCID: PMC527189
- DOI: 10.1105/tpc.104.021725
A gene regulatory network model for cell-fate determination during Arabidopsis thaliana flower development that is robust and recovers experimental gene expression profiles
Abstract
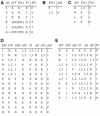
Flowers are icons in developmental studies of complex structures. The vast majority of 250,000 angiosperm plant species have flowers with a conserved organ plan bearing sepals, petals, stamens, and carpels in the center. The combinatorial model for the activity of the so-called ABC homeotic floral genes has guided extensive experimental studies in Arabidopsis thaliana and many other plant species. However, a mechanistic and dynamical explanation for the ABC model and prevalence among flowering plants is lacking. Here, we put forward a simple discrete model that postulates logical rules that formally summarize published ABC and non-ABC gene interaction data for Arabidopsis floral organ cell fate determination and integrates this data into a dynamic network model. This model shows that all possible initial conditions converge to few steady gene activity states that match gene expression profiles observed experimentally in primordial floral organ cells of wild-type and mutant plants. Therefore, the network proposed here provides a dynamical explanation for the ABC model and shows that precise signaling pathways are not required to restrain cell types to those found in Arabidopsis, but these are rather determined by the overall gene network dynamics. Furthermore, we performed robustness analyses that clearly show that the cell types recovered depend on the network architecture rather than on specific values of the model's gene interaction parameters. These results support the hypothesis that such a network constitutes a developmental module, and hence provide a possible explanation for the overall conservation of the ABC model and overall floral plan among angiosperms. In addition, we have been able to predict the effects of differences in network architecture between Arabidopsis and Petunia hybrida.
Figures




References
-
- Albert, R., Jeong, H., and Barabási, A.-L. (2000). Error and attack tolerance of complex networks. Nature 406, 378–382. - PubMed
-
- Ambrose, B.A., Lerner, D.R., Ciceri, P., Padilla, C.M., Yanofsky, M.F., and Schmidt, R.J. (2000). Molecular and genetic analyses of the Silky1 gene reveal conservation of floral organ specification between Eudicots and Monocots. Mol. Cell 5, 569–579. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

