Protein kinase CK2 phosphorylation regulates the interaction of Kaposi's sarcoma-associated herpesvirus regulatory protein ORF57 with its multifunctional partner hnRNP K
- PMID: 15486205
- PMCID: PMC524287
- DOI: 10.1093/nar/gkh876
Protein kinase CK2 phosphorylation regulates the interaction of Kaposi's sarcoma-associated herpesvirus regulatory protein ORF57 with its multifunctional partner hnRNP K
Retraction in
-
Retraction of 'Protein kinase CK2 phosphorylation regulates the interaction of Kaposi's sarcoma-associated herpesvirus regulatory protein ORF57 with its multifunctional partner hnRNP K'.Nucleic Acids Res. 2022 Jul 22;50(13):7800. doi: 10.1093/nar/gkac574. Nucleic Acids Res. 2022. PMID: 35786728 Free PMC article. No abstract available.
Expression of concern in
-
Editorial Expression of Concern on 'Protein kinase CK2 phosphorylation regulates the interaction of Kaposi's sarcoma-associated herpesvirus regulatory protein ORF57 with its multifunctional partner hnRNP K'.Nucleic Acids Res. 2022 Jan 25;50(2):1200. doi: 10.1093/nar/gkab1294. Nucleic Acids Res. 2022. PMID: 34951449 Free PMC article. No abstract available.
Abstract
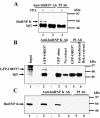
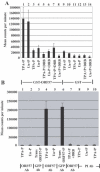
ORF57 protein of Kaposi's sarcoma-associated herpesvirus has a counterpart in all herpesvirus of mammals and birds and regulates gene expression at transcriptional and post-transcriptional levels. ORF57 was capable of self-interaction and bound a rapidly migrating form of heterogeneous nuclear ribonucleoprotein K (hnRNP K), a multifunctional cellular protein involved in gene expression. In virus infected cell extracts, ORF57 was present in a complex with hnRNP K that had protein kinase CK2 activity, and was phosphorylated by CK2. Different regions of ORF57 bound both catalytic alpha/alpha' and regulatory beta subunits of CK2. CK2 modification enhanced the ORF57-hnRNP K interaction, and may regulate the presence and activities of components in the complex. We suggest that ORF57 and hnRNP K interaction may modulate ORF57-mediated regulation of viral gene expression. Herpesviral ORF57 (Rhadinovirus) and ICP27 (Simplexvirus) proteins both interact with hnRNP K and CK2 implying that adaptation of the ancestral hnRNP K and CK2 to associate with viral regulatory ancestor protein likely pre-dates divergence of these Herpesviridae genera that occurred 200 million years ago.
Figures





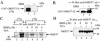

References
-
- Davison A.J. and Clements,J.B. (1997) Herpesviruses: general properties. In Mahy,B.W.J. and Collier,L.H. (eds), Topley & Wilson's Principles of Bacteriology, Virology and Immunology. 9th edn. Edward Arnold, London, pp. 309–323.
-
- Roizman B. and Knipe,D.M. (2001) In Knipe,D.M. and Howley,P.M. (eds), Fields Virology. 4th edn. Lippincott Williams & Wilkins, Philadelphia, Vol. 2, pp. 2399–2459.
-
- Chang Y., Cesarman,E., Pessin,M.S., Lee,F., Culpepper,J., Knowles,D.M. and Moore,P.S. (1994) Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science, 266, 1865–1869. - PubMed
-
- Viejo-Borbolla A. and Schulz,T.F. (2003) Kaposi's sarcoma-associated herpesvirus (KSHV/HHV8): key aspects of epidemiology and pathogenesis. AIDS Rev., 5, 222–229. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

