Diversity of guanylate cyclase-activating proteins (GCAPs) in teleost fish: characterization of three novel GCAPs (GCAP4, GCAP5, GCAP7) from zebrafish (Danio rerio) and prediction of eight GCAPs (GCAP1-8) in pufferfish (Fugu rubripes)
- PMID: 15486694
- PMCID: PMC1351297
- DOI: 10.1007/s00239-004-2614-y
Diversity of guanylate cyclase-activating proteins (GCAPs) in teleost fish: characterization of three novel GCAPs (GCAP4, GCAP5, GCAP7) from zebrafish (Danio rerio) and prediction of eight GCAPs (GCAP1-8) in pufferfish (Fugu rubripes)
Abstract
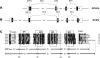
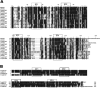
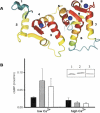
The guanylate cyclase-activating proteins (GCAPs) are Ca(2+)-binding proteins of the calmodulin (CaM) gene superfamily that function in the regulation of photoreceptor guanylate cyclases (GCs). In the mammalian retina, two GCAPs (GCAP 1-2) and two transmembrane GCs have been identified as part of a complex regulatory system responsive to fluctuating levels of free Ca(2+). A third GCAP, GCAP3, is expressed in human and zebrafish (Danio rerio) retinas, and a guanylate cyclase-inhibitory protein (GCIP) has been shown to be present in frog cones. To explore the diversity of GCAPs in more detail, we searched the pufferfish (Fugu rubripes) and zebrafish (Danio rerio) genomes for GCAP-related gene sequences (fuGCAPs and zGCAPs, respectively) and found that at least five additional GCAPs (GCAP4-8) are predicted to be present in these species. We identified genomic contigs encoding fuGCAPl-8, fuGCIP, zGCAPl-5, zGCAP7 and zGCIP. We describe cloning, expression and localization of three novel GCAPs present in the zebrafish retina (zGCAP4, zGCAP5, and zGCAP7). The results show that recombinant zGCAP4 stimulated bovine rod outer segment GC in a Ca(2+)-dependent manner. RT-PCR with zGCAP specific primers showed specific expression of zGCAPs and zGCIP in the retina, while zGCAPl mRNA is also present in the brain. In situ hybridization with anti-sense zGCAP4, zGCAP5 and zGCAP7 RNA showed exclusive expression in zebrafish cone photoreceptors. The presence of at least eight GCAP genes suggests an unexpected diversity within this subfamily of Ca(2+)-binding proteins in the teleost retina, and suggests additional functions for GCAPs apart from stimulation of GC. Based on genome searches and EST analyses, the mouse and human genomes do not harbor GCAP4-8 or GCIP genes.
Figures



References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Ames JB, Dizhoor AM, Ikura M, Palczewski K, Stryer L. Three-dimensional structure of guanylyl cyclase activating protein-2, a calcium-sensitive modulator of photoreceptor guanylyl cyclases. J Biol Chem. 1999;274:19329–19337. - PubMed
-
- Aparicio S, Chapman J, Stupka E, Putnam N, Chia JM, Dehal P, Christoffels A, Rash S, Hoon S, Smit A, Gelpke MD, Roach J, Oh T, Ho IY, Wong M, Detter C, Verhoef F, Predki P, Tay A, Lucas S, Richardson P, Smith SF, Clark MS, Edwards YJ, Doggett N, Zharkikh A, Tavtigian SV, Pruss D, Barnstead M, Evans C, Baden H, Powell J, Glusman G, Rowen L, Hood L, Tan YH, Elgar G, Hawkins T, Venkatesh B, Rokhsar D, Brenner S. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–1310. - PubMed
-
- Applebury ML, Antoch MP, Baxter LC, Chun LL, Falk JD, Farhangfar F, Kage K, Krzystolik MG, Lyass LA, Robbins JT. The murine cone photoreceptor: A single cone type expresses both S and M opsins with retinal spatial patterning. Neuron. 2000;27:513–523. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

