Kinetic investigation of human dipeptidyl peptidase II (DPPII)-mediated hydrolysis of dipeptide derivatives and its identification as quiescent cell proline dipeptidase (QPP)/dipeptidyl peptidase 7 (DPP7)
- PMID: 15487984
- PMCID: PMC1134796
- DOI: 10.1042/BJ20041156
Kinetic investigation of human dipeptidyl peptidase II (DPPII)-mediated hydrolysis of dipeptide derivatives and its identification as quiescent cell proline dipeptidase (QPP)/dipeptidyl peptidase 7 (DPP7)
Abstract
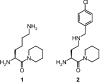
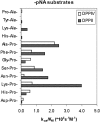
The presence of DPPII (dipeptidyl peptidase II; E.C. 3.4.14.2) has been demonstrated in various mammalian tissues. However, a profound molecular and catalytic characterization, including substrate selectivity, kinetics and pH-dependence, has not been conducted. In the present study, DPPII was purified from human seminal plasma to apparent homogeneity with a high yield (40%) purification scheme, including an inhibitor-based affinity chromatographic step. The inhibitor lysyl-piperidide (K(i) approximately 0.9 microM at pH 5.5) was chosen, as it provided a favourable affinity/recovery ratio. The human enzyme appeared as a 120 kDa homodimer. Mass spectrometric analysis after tryptic digestion together with a kinetic comparison indicate strongly its identity with QPP (quiescent cell proline dipeptidase), also called dipeptidyl peptidase 7. pH profiles of both kcat and kcat/K(m) clearly demonstrated that DPPII/QPP possesses an acidic and not a neutral optimum as was reported for QPP. Kinetic parameters of the human natural DPPII for dipeptide-derived chromogenic [pNA (p-nitroanilide)] and fluorogenic [4Me2NA (4-methoxy-2-naphthylamide)] substrates were determined under different assay conditions. DPPII preferred the chromogenic pNA-derived substrates over the fluorogenic 4Me2NA-derived substrates. Natural human DPPII showed high efficiency towards synthetic substrates containing proline at the P1 position and lysine at P2. The importance of the P1' group for P2 and P1 selectivity was revealed, explaining many discrepancies in the literature. Furthermore, substrate preferences of human DPPII and dipeptidyl peptidase IV were compared based on their selectivity constants (kcat/K(m)). Lys-Pro-pNA (k(cat)/K(m) 4.1x10(6) s(-1) x M(-1)) and Ala-Pro-pNA (kcat/K(m) 2.6x10(6) s(-1) x M(-1)) were found to be the most sensitive chromogenic substrates for human DPPII, but were less selective than Lys-Ala-pNA (kcat/K(m) 0.4x10(6) s(-1) x M(-1)).
Figures




Similar articles
-
Exploring the active site of tripeptidyl-peptidase II through studies of pH dependence of reaction kinetics.Biochim Biophys Acta. 2012 Apr;1824(4):561-70. doi: 10.1016/j.bbapap.2012.01.004. Epub 2012 Jan 14. Biochim Biophys Acta. 2012. PMID: 22266401
-
Dipeptidyl peptidases 8 and 9: specificity and molecular characterization compared with dipeptidyl peptidase IV.Biochem J. 2006 Jun 1;396(2):391-9. doi: 10.1042/BJ20060079. Biochem J. 2006. PMID: 16475979 Free PMC article.
-
Hydrolysis of dipeptide derivatives reveals the diversity in the M49 family.Biol Chem. 2013 Jun;394(6):767-71. doi: 10.1515/hsz-2012-0347. Biol Chem. 2013. PMID: 23362197
-
Dipeptidyl peptidase II (DPPII), a review.Clin Chim Acta. 2007 May 1;380(1-2):31-49. doi: 10.1016/j.cca.2007.01.024. Epub 2007 Feb 1. Clin Chim Acta. 2007. PMID: 17328877 Review.
-
Advances in understanding the expression and function of dipeptidyl peptidase 8 and 9.Mol Cancer Res. 2013 Dec;11(12):1487-96. doi: 10.1158/1541-7786.MCR-13-0272. Epub 2013 Sep 13. Mol Cancer Res. 2013. PMID: 24038034 Review.
Cited by
-
Identification of selective and potent inhibitors of fibroblast activation protein and prolyl oligopeptidase.J Med Chem. 2013 May 9;56(9):3467-77. doi: 10.1021/jm400351a. Epub 2013 Apr 29. J Med Chem. 2013. PMID: 23594271 Free PMC article.
-
Enzyme activity and immunohistochemical localization of dipeptidyl peptidase 8 and 9 in male reproductive tissues.J Histochem Cytochem. 2009 Jun;57(6):531-41. doi: 10.1369/jhc.2009.952739. Epub 2009 Feb 2. J Histochem Cytochem. 2009. PMID: 19188489 Free PMC article.
-
DPP8/DPP9 inhibition elicits canonical Nlrp1b inflammasome hallmarks in murine macrophages.Life Sci Alliance. 2019 Feb 4;2(1):e201900313. doi: 10.26508/lsa.201900313. Print 2019 Feb. Life Sci Alliance. 2019. PMID: 30718379 Free PMC article.
-
The Dipeptidyl Peptidase Family, Prolyl Oligopeptidase, and Prolyl Carboxypeptidase in the Immune System and Inflammatory Disease, Including Atherosclerosis.Front Immunol. 2015 Aug 7;6:387. doi: 10.3389/fimmu.2015.00387. eCollection 2015. Front Immunol. 2015. PMID: 26300881 Free PMC article. Review.
-
Structural and time-resolved mechanistic investigations of protein hydrolysis by the acidic proline-specific endoprotease from Aspergillus niger.Protein Sci. 2024 Jan;33(1):e4856. doi: 10.1002/pro.4856. Protein Sci. 2024. PMID: 38059672 Free PMC article.
References
-
- Park J. E., Lenter M. C., Zimmermann R. N., Garin-Chesa P., Old L. J., Rettig W. J. Fibroblast activation protein, a dual specificity serine protease expressed in reactive human tumor stromal fibroblasts. J. Biol. Chem. 1999;274:36505–36512. - PubMed
-
- Abbott C. A., Yu D. M., Woollatt E., Sutherland G. R., McCaughan G. W., Gorrell M. D. Cloning, expression and chromosomal localization of a novel human dipeptidyl peptidase (DPP) IV homolog, DPP8. Eur. J. Biochem. 2000;267:6140–6150. - PubMed
-
- Olsen C., Wagtmann N. Identification and characterization of human DPP9, a novel homologue of dipeptidyl peptidase IV. Gene. 2002;299:185–193. - PubMed
-
- Lambeir A. M., Durinx C., Scharpe S., De Meester I. Dipeptidyl-peptidase IV from bench to bedside: an update on structural properties, functions, and clinical aspects of the enzyme DPP IV. Crit. Rev. Clin. Lab. Sci. 2003;40:209–294. - PubMed
-
- McDonald J. K., Ellis S., Reilly T. J. Properties of dipeptidyl arylamidase I of the pituitary. J. Biol. Chem. 1966;241:1494–1501. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

