Caryoscope: an Open Source Java application for viewing microarray data in a genomic context
- PMID: 15488149
- PMCID: PMC528725
- DOI: 10.1186/1471-2105-5-151
Caryoscope: an Open Source Java application for viewing microarray data in a genomic context
Abstract
Background: Microarray-based comparative genome hybridization experiments generate data that can be mapped onto the genome. These data are interpreted more easily when represented graphically in a genomic context.
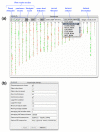
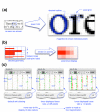
Results: We have developed Caryoscope, which is an open source Java application for visualizing microarray data from array comparative genome hybridization experiments in a genomic context. Caryoscope can read General Feature Format files (GFF files), as well as comma- and tab-delimited files, that define the genomic positions of the microarray reporters for which data are obtained. The microarray data can be browsed using an interactive, zoomable interface, which helps users identify regions of chromosomal deletion or amplification. The graphical representation of the data can be exported in a number of graphic formats, including publication-quality formats such as PostScript.
Conclusion: Caryoscope is a useful tool that can aid in the visualization, exploration and interpretation of microarray data in a genomic context.
Figures




References
-
- Pollack JR, Sorlie T, Perou CM, Rees CA, Jeffrey SS, Lonning PE, Tibshirani R, Botstein D, Borresen-Dale AL, Brown PO. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci U S A. 2002;99:12963–12968. doi: 10.1073/pnas.162471999. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

