Maximizing the efficacy of SAGE analysis identifies novel transcripts in Arabidopsis
- PMID: 15489285
- PMCID: PMC523381
- DOI: 10.1104/pp.104.043406
Maximizing the efficacy of SAGE analysis identifies novel transcripts in Arabidopsis
Abstract
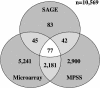
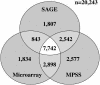
The efficacy of using Serial Analysis of Gene Expression (SAGE) to analyze the transcriptome of the model dicotyledonous plant Arabidopsis was assessed. We describe an iterative tag-to-gene matching process that exploits the availability of the whole genome sequence of Arabidopsis. The expression patterns of 98% of the annotated Arabidopsis genes could theoretically be evaluated through SAGE and using an iterative matching process 79% could be identified by a tag found at a unique site in the genome. A total of 145,170 reliable experimental tags from two Arabidopsis leaf tissue SAGE libraries were analyzed, of which 29,632 were distinct. The majority (93%) of the 12,988 experimental tags observed greater than once could be matched within the Arabidopsis genome. However, only 78% were matched to a single locus within the genome, reflecting the complexities associated with working in a highly duplicated genome. In addition to a comprehensive assessment of gene expression in Arabidopsis leaf tissue, we describe evidence of transcription from pseudo-genes as well as evidence of alternative mRNA processing and anti-sense transcription. This collection of experimental SAGE tags could be exploited to assist in the on-going annotation of the Arabidopsis genome.
Figures

References
-
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 - PubMed
-
- Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo S, McCurdy S, Foy M, Ewan M, et al (2000) Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotechnol 18: 630–634 - PubMed
-
- Carpousis AJ, Vanzo NF, Raynal LC (1999) mRNA degradation. A tale of poly(A) and multiprotein machines. Trends Genet 15: 24–28 - PubMed
-
- Cock JM, Swarup R, Dumas C (1997) Natural antisense transcripts of the S locus receptor kinase gene and related sequences in Brassica oleracea. Mol Gen Genet 255: 514–524 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

