The geminivirus nuclear shuttle protein is a virulence factor that suppresses transmembrane receptor kinase activity
- PMID: 15489295
- PMCID: PMC529541
- DOI: 10.1101/gad.1245904
The geminivirus nuclear shuttle protein is a virulence factor that suppresses transmembrane receptor kinase activity
Abstract
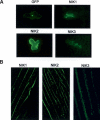
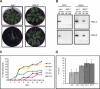
Despite the large number of leucine-rich-repeat (LRR) receptor-like-kinases (RLKs) in plants and their conceptual relevance in signaling events, functional information is restricted to a few family members. Here we describe the characterization of new LRR-RLK family members as virulence targets of the geminivirus nuclear shuttle protein (NSP). NSP interacts specifically with three LRR-RLKs, NIK1, NIK2, and NIK3, through an 80-amino acid region that encompasses the kinase active site and A-loop. We demonstrate that these NSP-interacting kinases (NIKs) are membrane-localized proteins with biochemical properties of signaling receptors. They behave as authentic kinase proteins that undergo autophosphorylation and can also phosphorylate exogenous substrates. Autophosphorylation occurs via an intermolecular event and oligomerization precedes the activation of the kinase. Binding of NSP to NIK inhibits its kinase activity in vitro, suggesting that NIK is involved in antiviral defense response. In support of this, infectivity assays showed a positive correlation between infection rate and loss of NIK1 and NIK3 function. Our data are consistent with a model in which NSP acts as a virulence factor to suppress NIK-mediated antiviral responses.
Figures




References
-
- Alonso J.M., Stepanova, A.N., Leisse, T.J., Kim, C.J., Chen, H., Shinn, P., Stevenson, D.K., Zimmerman, J., Barajas, P., Cheuk, R., et al. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653-657. - PubMed
-
- The Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796-815. - PubMed
-
- Cascardo J.C.M., Almeida, R.S., Buzeli, R.A.A., Carolino, S.M.B., Otoni, W.C., and Fontes, E.P.B. 2000. The phosphorylation state and expression of soybean BiP isoforms are differentially regulated following abiotic stresses. J. Biol. Chem. 275: 14494-14500. - PubMed
-
- Clark S.E., Williams, R.W., and Meyerowitz, E.M. 1997. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89: 575-585. - PubMed
-
- Dievart A. and Clark, S.E. 2004. LRR-containing receptors regulating plant development and defense. Development 131: 251-261. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
