The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis
- PMID: 15489296
- PMCID: PMC529542
- DOI: 10.1101/gad.317504
The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis
Abstract
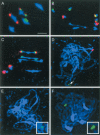
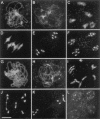
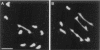
MSH4, a meiosis-specific member of the MutS-homolog family of genes, is required for normal levels of recombination and fertility in budding yeast, mouse, and Caenorhabditis elegans. In this paper, we report the identification and characterization of the Arabidopsis homolog of MSH4 (AtMSH4). We demonstrate that AtMSH4 expression can only be detected in floral tissues, consistent with a role in reproduction. Immunofluorescence studies indicate that its expression is limited to early meiotic prophase I, preceding the synapsis of homologous chromosomes. A T-DNA insertional mutant (Atmsh4) exhibited normal vegetative growth but a severe reduction in fertility, consistent with a meiotic defect; this was confirmed by cytological analysis of meiosis. RNAi-induced down-regulation of the MSH4 gene resulted in a similar fertility and meiotic phenotype. We demonstrate that prophase I chromosome synapsis is delayed and may be incomplete in Atmsh4, and metaphase I chiasma frequency is greatly reduced to approximately 15% of wild type, leading to univalence and nondisjunction. We show that these residual chiasmata are randomly distributed among cells and chromosomes. These features of chiasma frequency and distribution in Atmsh4 show close parallels to MSH4-independent crossovers in budding yeast that have been proposed to originate by a separate pathway. Furthermore, the characteristics of the MSH4-independent chiasmata in the Atmsh4 mutant closely parallel those of second-pathway crossovers that have been postulated from Arabidopsis crossover analysis and mathematical modeling. Taken together, this evidence strongly indicates that Arabidopsis possesses two crossover pathways.
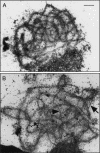
Figures





Similar articles
-
Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over.Plant J. 2008 Apr;54(1):152-62. doi: 10.1111/j.1365-313X.2008.03403.x. Epub 2008 Jan 7. Plant J. 2008. PMID: 18182028
-
AtMSH5 partners AtMSH4 in the class I meiotic crossover pathway in Arabidopsis thaliana, but is not required for synapsis.Plant J. 2008 Jul;55(1):28-39. doi: 10.1111/j.1365-313X.2008.03470.x. Plant J. 2008. PMID: 18318687
-
The Arabidopsis MutS homolog AtMSH5 is required for normal meiosis.Cell Res. 2008 May;18(5):589-99. doi: 10.1038/cr.2008.44. Cell Res. 2008. PMID: 18379590
-
Meiotic cytology and chromosome behaviour in wild-type Arabidopsis thaliana.J Exp Bot. 2003 Jan;54(380):1-10. doi: 10.1093/jxb/erg034. J Exp Bot. 2003. PMID: 12456750 Review.
-
ASY1 coordinates early events in the plant meiotic recombination pathway.Cytogenet Genome Res. 2008;120(3-4):302-12. doi: 10.1159/000121079. Epub 2008 May 23. Cytogenet Genome Res. 2008. PMID: 18504359 Review.
Cited by
-
Analysis of Arabidopsis genome-wide variations before and after meiosis and meiotic recombination by resequencing Landsberg erecta and all four products of a single meiosis.Genome Res. 2012 Mar;22(3):508-18. doi: 10.1101/gr.127522.111. Epub 2011 Nov 21. Genome Res. 2012. PMID: 22106370 Free PMC article.
-
DNA cytosine methyltransferases differentially regulate genome-wide hypermutation and interhomolog recombination in Trichoderma reesei meiosis.Nucleic Acids Res. 2024 Sep 9;52(16):9551-9573. doi: 10.1093/nar/gkae611. Nucleic Acids Res. 2024. PMID: 39021337 Free PMC article.
-
The Number of Meiotic Double-Strand Breaks Influences Crossover Distribution in Arabidopsis.Plant Cell. 2018 Oct;30(10):2628-2638. doi: 10.1105/tpc.18.00531. Epub 2018 Oct 3. Plant Cell. 2018. PMID: 30282794 Free PMC article.
-
Dissecting meiosis of rye using translational proteomics.Ann Bot. 2008 Apr;101(6):873-80. doi: 10.1093/aob/mcm202. Epub 2007 Aug 31. Ann Bot. 2008. PMID: 17766846 Free PMC article.
-
Why do plants need the ZMM crossover pathway? A snapshot of meiotic recombination from the perspective of interhomolog polymorphism.Plant Reprod. 2023 Mar;36(1):43-54. doi: 10.1007/s00497-022-00446-3. Epub 2022 Jul 12. Plant Reprod. 2023. PMID: 35819509 Free PMC article.
References
-
- Albini S.M. 1994. A karyotype of the Arabidopsis thaliana genome derived from synaptonemal complex analysis at prophase I of meiosis. Plant J. 5: 665-672.
-
- Albini S.M. and Jones, G.H. 1987. Synaptonemal complex spreading in Allium cepa and A. fistulosum. I. The initiation and sequence of pairing. Chromosoma 95: 324-338.
-
- Allers T. and Lichten, M. 2001. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106: 47-57. - PubMed
-
- Ariyoshi M., Vassylyev, D.G., Iwasaki, H., Nakamura, H., Shinagawa, H., and Morikawa, K. 1994. Atomic structure of the RuvC resolvase: A Holliday junction-specific endonuclease from E. coli. Cell 78: 1063-1072. - PubMed
-
- Armstrong S.J. and Jones, G.H. 2001. Female meiosis in wildtype Arabidopsis thaliana and in two meiotic mutants. Sex Plant Reprod. 13: 177-183.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
