Systematic recovery and analysis of full-ORF human cDNA clones
- PMID: 15489330
- PMCID: PMC528924
- DOI: 10.1101/gr.2473704
Systematic recovery and analysis of full-ORF human cDNA clones
Abstract
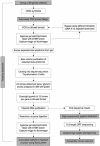
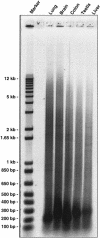
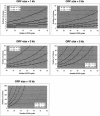
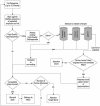
The Mammalian Gene Collection (MGC) consortium (http://mgc.nci.nih.gov) seeks to establish publicly available collections of full-ORF cDNAs for several organisms of significance to biomedical research, including human. To date over 15,200 human cDNA clones containing full-length open reading frames (ORFs) have been identified via systematic expressed sequence tag (EST) analysis of a diverse set of cDNA libraries; however, further systematic EST analysis is no longer an efficient method for identifying new cDNAs. As part of our involvement in the MGC program, we have developed a scalable method for targeted recovery of cDNA clones to facilitate recovery of genes absent from the MGC collection. First, cDNA is synthesized from various RNAs, followed by polymerase chain reaction (PCR) amplification of transcripts in 96-well plates using gene-specific primer pairs flanking the ORFs. Amplicons are cloned into a sequencing vector, and full-length sequences are obtained. Sequences are processed and assembled using Phred and Phrap, and analyzed using Consed and a number of bioinformatics methods we have developed. Sequences are compared with the Reference Sequence (RefSeq) database, and validation of sequence discrepancies is attempted using other sequence databases including dbEST and dbSNP. Clones with identical sequence to RefSeq or containing only validated changes will become part of the MGC human gene collection. Clones containing novel splice variants or polymorphisms have also been identified. Our approach to clone recovery, applied at large scale, has the potential to recover many and possibly most of the genes absent from the MGC collection.
Figures





References
-
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403-410. - PubMed
-
- Boguski, M.S., Lowe, T.M., and Tolstoshev, C.M. 1993. dbEST—Database for “expressed sequence tags”. Nat. Genet. 4: 332-333. - PubMed
-
- Butterfield, Y.S., Marra, M.A., Asano, J.K., Chan, S.Y., Guin, R., Krzywinski, M.I., Lee, S.S., MacDonald, K.W., Mathewson, C.A., Olson, T.E., et al. 2002. An efficient strategy for large-scale high-throughput transposon-mediated sequencing of cDNA clones. Nucleic Acids Res. 30: 2460-2468. - PMC - PubMed
WEB SITE REFERENCES
-
- http://genome.ucsc.edu/cgi-bin/hgBlat; Human BLAT Search.
-
- http://mgc.nci.nih.gov; Mammalian Gene Collection.
-
- http://www.ensembl.org; Ensembl.
-
- http://www.ncbi.nlm.nih.gov/dbEST; Expressed Sequence Tags database.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
