Concerted assembly and cloning of multiple DNA segments using in vitro site-specific recombination: functional analysis of multi-segment expression clones
- PMID: 15489333
- PMCID: PMC528927
- DOI: 10.1101/gr.2512204
Concerted assembly and cloning of multiple DNA segments using in vitro site-specific recombination: functional analysis of multi-segment expression clones
Abstract
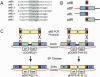
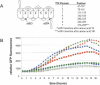
The ability to clone and manipulate DNA segments is central to molecular methods that enable expression, screening, and functional characterization of genes, proteins, and regulatory elements. We previously described the development of a novel technology that utilizes in vitro site-specific recombination to provide a robust and flexible platform for high-throughput cloning and transfer of DNA segments. By using an expanded repertoire of recombination sites with unique specificities, we have extended the technology to enable the high-efficiency in vitro assembly and concerted cloning of multiple DNA segments into a vector backbone in a predefined order, orientation, and reading frame. The efficiency and flexibility of this approach enables collections of functional elements to be generated and mixed in a combinatorial fashion for the parallel assembly of numerous multi-segment constructs. The assembled constructs can be further manipulated by directing exchange of defined segments with alternate DNA segments. In this report, we demonstrate feasibility of the technology and application to the generation of fusion proteins, the linkage of promoters to genes, and the assembly of multiple protein domains. The technology has broad implications for cell and protein engineering, the expression of multidomain proteins, and gene function analysis.
Figures
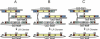




References
-
- Barak, L.S., Ferguson, S.S., Zhang, J., and Caron, M.G. 1997. A β-arrestin/green fluorescent protein biosensor for detecting G protein-coupled receptor activation. J. Biol. Chem. 272: 27497-27500. - PubMed
-
- Bardwell, L., Cook, J.G., Inouye, C.J., and Thorner, J. 1994. Signal propagation and regulation in the mating pheromone response pathway of the yeast Saccharomyces cerevisiae. Dev. Biol. 166: 363-379. - PubMed
-
- Bernard, P. and Couturier, M. 1992. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J. Mol. Biol. 226: 735-745. - PubMed
-
- Boulton, S.J., Gartner, A., Reboul, J., Vaglio, P., Dyson, N., Hill, D.E., and Vidal, M. 2002. Combined functional genomic maps of the C. elegans DNA damage response. Science 295: 127-131. - PubMed
WEB SITE REFERENCES
-
- http://www.fruitfly.org/EST/gateway.shtml; Drosophila ORFs.
-
- http://xgc.nci.nih.gov; Xenopus ORFs.
-
- http://zgc.nci.nih.gov; zebrafish ORFs.
-
- http://mgc.nci.nih.gov; human, mouse and rat ORFs.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
