Lactobacillus plantarum bacteriophage LP65: a new member of the SPO1-like genus of the family Myoviridae
- PMID: 15489418
- PMCID: PMC523202
- DOI: 10.1128/JB.186.21.7069-7083.2004
Lactobacillus plantarum bacteriophage LP65: a new member of the SPO1-like genus of the family Myoviridae
Abstract
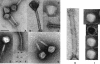
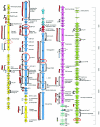
The virulent Lactobacillus plantarum myophage LP65 was isolated from industrial meat fermentation. Tail contraction led to reorganization of the tail sheath and the baseplate; a tail tube was extruded. In ultrathin section the phage adsorbed via its baseplate to the exterior of the cell, while the tail tube tunneled through the thick bacterial cell wall. Convoluted membrane structures were induced in the infected cell. Progeny phage was detected 100 min postinfection, and lysis occurred after extensive digestion of the cell wall. Sequence analysis revealed a genome of 131,573 bp of nonredundant DNA. Four major genome regions and a large tRNA gene cluster were observed. One module corresponded to DNA replication genes. Helicase/primase and two replication/recombination enzymes represented the only links to T4-like Myoviridae from gram-negative bacteria. Another module corresponded to the structural genes. Sequence relatedness identified links with Listeria phage A511, Staphylococcus phage K, and Bacillus phage SPO1. LP65 structural proteins were identified by two-dimensional proteome analysis and mass spectrometry. The putative tail sheath protein showed a shear-induced change in electrophoretic migration behavior. The genome organization of the structural module in LP65 resembled that of Siphoviridae from the lambda supergroup.
Figures





Comment in
-
Phage taxonomy: we agree to disagree.J Bacteriol. 2004 Nov;186(21):7029-31. doi: 10.1128/JB.186.21.7029-7031.2004. J Bacteriol. 2004. PMID: 15489416 Free PMC article. No abstract available.
References
-
- Altermann, E., J. R. Klein, and B. Henrich. 1999. Primary structure and features of the genome of the Lactobacillus gasseri temperate bacteriophage φadh. Gene 236:333-346. - PubMed
-
- Arisaka, F., J. Engel, and H. Klump. 1981. Contraction and dissociation of the bacteriophage T4 tail sheath induced by heat and urea. Prog. Clin. Biol. Res. 64:365-379. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

