The genome sequence of Mycoplasma hyopneumoniae strain 232, the agent of swine mycoplasmosis
- PMID: 15489423
- PMCID: PMC523201
- DOI: 10.1128/JB.186.21.7123-7133.2004
The genome sequence of Mycoplasma hyopneumoniae strain 232, the agent of swine mycoplasmosis
Abstract
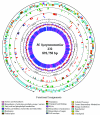
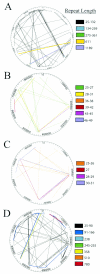
We present the complete genome sequence of Mycoplasma hyopneumoniae, an important member of the porcine respiratory disease complex. The genome is composed of 892,758 bp and has an average G+C content of 28.6 mol%. There are 692 predicted protein coding sequences, the average protein size is 388 amino acids, and the mean coding density is 91%. Functions have been assigned to 304 (44%) of the predicted protein coding sequences, while 261 (38%) of the proteins are conserved hypothetical proteins and 127 (18%) are unique hypothetical proteins. There is a single 16S-23S rRNA operon, and there are 30 tRNA coding sequences. The cilium adhesin gene has six paralogs in the genome, only one of which contains the cilium binding site. The companion gene, P102, also has six paralogs. Gene families constitute 26.3% of the total coding sequences, and the largest family is the 34-member ABC transporter family. Protein secretion occurs through a truncated pathway consisting of SecA, SecY, SecD, PrsA, DnaK, Tig, and LepA. Some highly conserved eubacterial proteins, such as GroEL and GroES, are notably absent. The DnaK-DnaJ-GrpR complex is intact, providing the only control over protein folding. There are several proteases that might serve as virulence factors, and there are 53 coding sequences with prokaryotic lipoprotein lipid attachment sites. Unlike other mycoplasmas, M. hyopneumoniae contains few genes with tandem repeat sequences that could be involved in phase switching or antigenic variation. Thus, it is not clear how M. hyopneumoniae evades the immune response and establishes a chronic infection.
Figures

References
-
- Artiushin, S., M. Duvall, and F. C. Minion. 1995. Phylogenetic analysis of mycoplasma strain ISM1499 and its assignment to the Acholeplasma oculi strain cluster. Int. J. Syst. Bacteriol. 45:104-109. - PubMed
-
- Bereiter, M., T. F. Young, H. S. Joo, and R. F. Ross. 1990. Evaluation of the ELISA, and comparison to the complement fixation test and radial immunodiffusion enzyme assay for detection of antibodies against Mycoplasma hyopneumoniae in swine serum. Vet. Microbiol. 25:177-192. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

