MorA defines a new class of regulators affecting flagellar development and biofilm formation in diverse Pseudomonas species
- PMID: 15489433
- PMCID: PMC523210
- DOI: 10.1128/JB.186.21.7221-7228.2004
MorA defines a new class of regulators affecting flagellar development and biofilm formation in diverse Pseudomonas species
Abstract
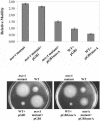
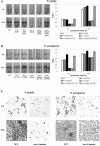
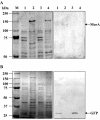
Assembly of bacterial flagella is developmentally important during both planktonic cell growth and biofilm formation. Flagellar biogenesis is complex, requiring coordinated expression of over 40 genes, and normally commences during the log-to-stationary transition phase. We describe here a novel membrane-localized regulator, MorA, that controls the timing of flagellar development and affects motility, chemotaxis, and biofilm formation in Pseudomonas putida. MorA is conserved among diverse Pseudomonas species, and homologues are present in all Pseudomonas genomes sequenced thus far. In P. putida, the absence of MorA derepresses flagellar development, which leads to constitutive formation of flagella in the mutant cells in all growth phases. In Pseudomonas aeruginosa, the absence of MorA led to a reduction in biofilm formation. However, unlike the motility of P. putida, the motility of the P. aeruginosa mutants was unaffected. Our data illustrate a novel developmentally regulated sensory and signaling pathway for several properties required for virulence and ecological fitness of Pseudomonas species.
Figures



References
-
- Adaikkalam, V., and S. Swarup. 2002. Molecular characterization of an operon, cueAR, encoding a putative P1-type ATPase and a MerR-type regulatory protein involved in copper homeostasis in Pseudomonas putida. Microbiology 148:2857-2867. - PubMed
-
- Ausmees, N., R. Mayer, H. Weinhouse, G. Volman, D. Amikam, M. Benziman, and M. Lindberg. 2001. Genetic data indicate that proteins containing the GGDEF domain possess diguanylate cyclase activity. FEMS Microbiol. Lett. 204:163-167. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

