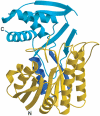
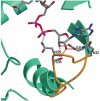
Crystal structure of Escherichia coli ArnA (PmrI) decarboxylase domain. A key enzyme for lipid A modification with 4-amino-4-deoxy-L-arabinose and polymyxin resistance
- PMID: 15491143
- PMCID: PMC2680612
- DOI: 10.1021/bi048551f
Crystal structure of Escherichia coli ArnA (PmrI) decarboxylase domain. A key enzyme for lipid A modification with 4-amino-4-deoxy-L-arabinose and polymyxin resistance
Abstract
Gram-negative bacteria including Escherichia coli, Salmonella typhimurium, and Pseudomonas aeruginosa can modify the structure of lipid A in their outer membrane with 4-amino-4-deoxy-l-arabinose (Ara4N). Such modification results in resistance to cationic antimicrobial peptides of the innate immune system and antibiotics such as polymyxin. ArnA is a key enzyme in the lipid A modification pathway, and its deletion abolishes both the Ara4N-lipid A modification and polymyxin resistance. ArnA is a bifunctional enzyme. It can catalyze (i) the NAD(+)-dependent decarboxylation of UDP-glucuronic acid to UDP-4-keto-arabinose and (ii) the N-10-formyltetrahydrofolate-dependent formylation of UDP-4-amino-4-deoxy-l-arabinose. We show that the NAD(+)-dependent decarboxylating activity is contained in the 360 amino acid C-terminal domain of ArnA. This domain is separable from the N-terminal fragment, and its activity is identical to that of the full-length enzyme. The crystal structure of the ArnA decarboxylase domain from E. coli is presented here. The structure confirms that the enzyme belongs to the short-chain dehydrogenase/reductase (SDR) family. On the basis of sequence and structure comparisons of the ArnA decarboxylase domain with other members of the short-chain dehydrogenase/reductase (SDR) family, we propose a binding model for NAD(+) and UDP-glucuronic acid and the involvement of residues T(432), Y(463), K(467), R(619), and S(433) in the mechanism of NAD(+)-dependent oxidation of the 4''-OH of the UDP-glucuronic acid and decarboxylation of the UDP-4-keto-glucuronic acid intermediate.
Figures






Similar articles
-
A formyltransferase required for polymyxin resistance in Escherichia coli and the modification of lipid A with 4-Amino-4-deoxy-L-arabinose. Identification and function oF UDP-4-deoxy-4-formamido-L-arabinose.J Biol Chem. 2005 Apr 8;280(14):14154-67. doi: 10.1074/jbc.M414265200. Epub 2005 Jan 28. J Biol Chem. 2005. PMID: 15695810
-
Crystal structure and mechanism of the Escherichia coli ArnA (PmrI) transformylase domain. An enzyme for lipid A modification with 4-amino-4-deoxy-L-arabinose and polymyxin resistance.Biochemistry. 2005 Apr 12;44(14):5328-38. doi: 10.1021/bi047384g. Biochemistry. 2005. PMID: 15807526 Free PMC article.
-
Oxidative decarboxylation of UDP-glucuronic acid in extracts of polymyxin-resistant Escherichia coli. Origin of lipid a species modified with 4-amino-4-deoxy-L-arabinose.J Biol Chem. 2002 Jan 25;277(4):2886-96. doi: 10.1074/jbc.M109377200. Epub 2001 Nov 8. J Biol Chem. 2002. PMID: 11706007
-
UDP-hexose 4-epimerases: a view on structure, mechanism and substrate specificity.Carbohydr Res. 2015 Sep 23;414:8-14. doi: 10.1016/j.carres.2015.06.006. Epub 2015 Jun 21. Carbohydr Res. 2015. PMID: 26162744 Review.
-
Stereo-electronic control of reaction selectivity in short-chain dehydrogenases: Decarboxylation, epimerization, and dehydration.Curr Opin Chem Biol. 2021 Apr;61:43-52. doi: 10.1016/j.cbpa.2020.09.010. Epub 2020 Nov 6. Curr Opin Chem Biol. 2021. PMID: 33166830 Review.
Cited by
-
Structure and mechanism of ArnA: conformational change implies ordered dehydrogenase mechanism in key enzyme for polymyxin resistance.Structure. 2005 Jun;13(6):929-42. doi: 10.1016/j.str.2005.03.018. Structure. 2005. PMID: 15939024 Free PMC article.
-
Resistance to antimicrobial peptides and stress response in Mycoplasma pulmonis.Antimicrob Agents Chemother. 2005 Oct;49(10):4154-65. doi: 10.1128/AAC.49.10.4154-4165.2005. Antimicrob Agents Chemother. 2005. PMID: 16189093 Free PMC article.
-
Different surface charge of colistin-susceptible and -resistant Acinetobacter baumannii cells measured with zeta potential as a function of growth phase and colistin treatment.J Antimicrob Chemother. 2011 Jan;66(1):126-33. doi: 10.1093/jac/dkq422. Epub 2010 Nov 16. J Antimicrob Chemother. 2011. PMID: 21081544 Free PMC article.
-
Synthesis of and evaluation of lipid A modification by 4-substituted 4-deoxy arabinose analogs as potential inhibitors of bacterial polymyxin resistance.Bioorg Med Chem Lett. 2008 Feb 15;18(4):1507-10. doi: 10.1016/j.bmcl.2007.12.061. Epub 2007 Dec 27. Bioorg Med Chem Lett. 2008. PMID: 18187325 Free PMC article.
-
Single-Molecule Resolution of Antimicrobial Peptide Interactions with Supported Lipid A Bilayers.Biophys J. 2018 Jun 5;114(11):2606-2616. doi: 10.1016/j.bpj.2018.04.019. Biophys J. 2018. PMID: 29874611 Free PMC article.
References
-
- Hoffmann JA, Kafatos FC, Janeway CA, Ezekowitz RA. Phylogenetic perspectives in innate immunity. Science. 1999;284:1313–1318. - PubMed
-
- Scott MG, Hancock RE. Cationic antimicrobial peptides and their multifunctional role in the immune system. Crit. Rev. Immunol. 2000;20:407–431. - PubMed
-
- Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. - PubMed
-
- Zasloff M. Antibiotic peptides as mediators of innate immunity. Curr. Opin. Immunol. 1992;4:3–7. - PubMed
-
- Gunn JS, Ryan SS, Van Velkinburgh JC, Ernst RK, Miller SI. Genetic and functional analysis of a PmrA-PmrB-regulated locus necessary for lipopolysaccharide modification, antimicrobial peptide resistance, and oral virulence of Salmonella enterica serovar typhimurium. Infect Immun. 2000;68:6139–6146. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

