Global position and recruitment of HATs and HDACs in the yeast genome
- PMID: 15494307
- PMCID: PMC3004369
- DOI: 10.1016/j.molcel.2004.09.021
Global position and recruitment of HATs and HDACs in the yeast genome
Abstract
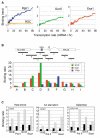
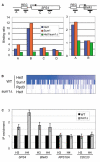
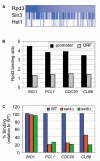
Chromatin regulators play fundamental roles in the regulation of gene expression and chromosome maintenance, but the regions of the genome where most of these regulators function has not been established. We explored the genome-wide occupancy of four different chromatin regulators encoded in Saccharomyces cerevisiae. The results reveal that the histone acetyltransferases Gcn5 and Esa1 are both generally recruited to the promoters of active protein-coding genes. In contrast, the histone deacetylases Hst1 and Rpd3 are recruited to specific sets of genes associated with distinct cellular functions. Our results provide new insights into the association of histone acetyltransferases and histone deacetylases with the yeast genome, and together with previous studies, suggest how these chromatin regulators are recruited to specific regions of the genome.
Figures

References
-
- Baek SH, Ohgi KA, Rose DW, Koo EH, Glass CK, Rosenfeld MG. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-kappaB and beta-amyloid precursor protein. Cell. 2002;110:55–67. - PubMed
-
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

