A conserved transcript pattern in response to a specialist and a generalist herbivore
- PMID: 15494554
- PMCID: PMC527203
- DOI: 10.1105/tpc.104.026120
A conserved transcript pattern in response to a specialist and a generalist herbivore
Abstract
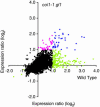
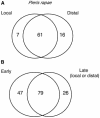
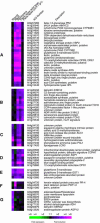
Transcript patterns elicited in response to attack reveal, at the molecular level, how plants respond to aggressors. These patterns are fashioned both by inflicted physical damage as well as by biological components displayed or released by the attacker. Different types of attacking organisms might therefore be expected to elicit different transcription programs in the host. Using a large-scale DNA microarray, we characterized gene expression in damaged as well as in distal Arabidopsis thaliana leaves in response to the specialist insect, Pieris rapae. More than 100 insect-responsive genes potentially involved in defense were identified, including genes involved in pathogenesis, indole glucosinolate metabolism, detoxification and cell survival, and signal transduction. Of these 114 genes, 111 were induced in Pieris feeding, and only three were repressed. Expression patterns in distal leaves were markedly similar to those of local leaves. Analysis of wild-type and jasmonate mutant plants, coupled with jasmonate treatment, showed that between 67 and 84% of Pieris-regulated gene expression was controlled, totally or in part, by the jasmonate pathway. This was correlated with increased larval performance on the coronatine insensitive1 glabrous1 (coi1-1 gl1) mutant. Independent mutations in COI1 and GL1 led to a faster larval weight gain, but the gl1 mutation had relatively little effect on the expression of the insect-responsive genes examined. Finally, we compared transcript patterns in Arabidopis in response to larvae of the specialist P. rapae and to a generalist insect, Spodoptera littoralis. Surprisingly, given the complex nature of insect salivary components and reported differences between species, almost identical transcript profiles were observed. This study also provides a robustly characterized gene set for the further investigation of plant-insect interaction.
Figures





References
-
- Agrawal, A.A. (1998). Induced responses to herbivory and increased plant performance. Science 279, 1201–1202. - PubMed
-
- Agrawal, A.A. (1999). Induced responses to herbivory in wild radish: Effects on several herbivores and plant fitness. Ecology 80, 1713–1723.
-
- Agrawal, A.A., and Kurashige, N.S. (2003). A role for isothiocyanates in plant resistance against the specialist herbivore Pieris rapae. J. Chem. Ecol. 29, 1403–1415. - PubMed
-
- Alborn, H.T., Brennan, M.M., and Tumlinson, J.H. (2003). Differential activity and degradation of plant volatile elicitors in regurgitant of tobacco hornworm (Manduca sexta) larvae. J. Chem. Ecol. 29, 1357–1372. - PubMed
-
- Alborn, H.T., Turlings, T.C.J., Jones, T.H., Stenhagen, G., Loughrin, J.H., and Tumlinson, J.H. (1997). An elicitor of plant volatiles from beet armyworm oral secretion. Science 276, 945–949.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

