Phylogenetic inference in Rafflesiales: the influence of rate heterogeneity and horizontal gene transfer
- PMID: 15496229
- PMCID: PMC528834
- DOI: 10.1186/1471-2148-4-40
Phylogenetic inference in Rafflesiales: the influence of rate heterogeneity and horizontal gene transfer
Abstract
Background: The phylogenetic relationships among the holoparasites of Rafflesiales have remained enigmatic for over a century. Recent molecular phylogenetic studies using the mitochondrial matR gene placed Rafflesia, Rhizanthes and Sapria (Rafflesiaceae s. str.) in the angiosperm order Malpighiales and Mitrastema (Mitrastemonaceae) in Ericales. These phylogenetic studies did not, however, sample two additional groups traditionally classified within Rafflesiales (Apodantheaceae and Cytinaceae). Here we provide molecular phylogenetic evidence using DNA sequence data from mitochondrial and nuclear genes for representatives of all genera in Rafflesiales.
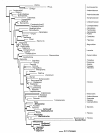
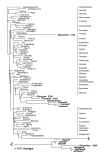
Results: Our analyses indicate that the phylogenetic affinities of the large-flowered clade and Mitrastema, ascertained using mitochondrial matR, are congruent with results from nuclear SSU rDNA when these data are analyzed using maximum likelihood and Bayesian methods. The relationship of Cytinaceae to Malvales was recovered in all analyses. Relationships between Apodanthaceae and photosynthetic angiosperms varied depending upon the data partition: Malvales (3-gene), Cucurbitales (matR) or Fabales (atp1). The latter incongruencies suggest that horizontal gene transfer (HGT) may be affecting the mitochondrial gene topologies. The lack of association between Mitrastema and Ericales using atp1 is suggestive of HGT, but greater sampling within eudicots is needed to test this hypothesis further.
Conclusions: Rafflesiales are not monophyletic but composed of three or four independent lineages (families): Rafflesiaceae, Mitrastemonaceae, Apodanthaceae and Cytinaceae. Long-branch attraction appears to be misleading parsimony analyses of nuclear small-subunit rDNA data, but model-based methods (maximum likelihood and Bayesian analyses) recover a topology that is congruent with the mitochondrial matR gene tree, thus providing compelling evidence for organismal relationships. Horizontal gene transfer appears to be influencing only some taxa and some mitochondrial genes, thus indicating that the process is acting at the single gene (not whole genome) level.
Figures



Similar articles
-
Increased sampling of both genes and taxa improves resolution of phylogenetic relationships within Magnoliidae, a large and early-diverging clade of angiosperms.Mol Phylogenet Evol. 2014 Jan;70:84-93. doi: 10.1016/j.ympev.2013.09.010. Epub 2013 Sep 18. Mol Phylogenet Evol. 2014. PMID: 24055602
-
Mitochondrial matR sequences help to resolve deep phylogenetic relationships in rosids.BMC Evol Biol. 2007 Nov 10;7:217. doi: 10.1186/1471-2148-7-217. BMC Evol Biol. 2007. PMID: 17996110 Free PMC article.
-
Host-to-parasite gene transfer in flowering plants: phylogenetic evidence from Malpighiales.Science. 2004 Jul 30;305(5684):676-8. doi: 10.1126/science.1100671. Epub 2004 Jul 15. Science. 2004. PMID: 15256617
-
Mitochondria in parasitic plants.Mitochondrion. 2020 May;52:173-182. doi: 10.1016/j.mito.2020.03.008. Epub 2020 Mar 26. Mitochondrion. 2020. PMID: 32224234 Review.
-
Horizontal Gene Transfer Involving Chloroplasts.Int J Mol Sci. 2021 Apr 25;22(9):4484. doi: 10.3390/ijms22094484. Int J Mol Sci. 2021. PMID: 33923118 Free PMC article. Review.
Cited by
-
Genomic reconfiguration in parasitic plants involves considerable gene losses alongside global genome size inflation and gene births.Plant Physiol. 2021 Jul 6;186(3):1412-1423. doi: 10.1093/plphys/kiab192. Plant Physiol. 2021. PMID: 33909907 Free PMC article.
-
Life history, diversity, and distribution in parasitic flowering plants.Plant Physiol. 2021 Sep 4;187(1):32-51. doi: 10.1093/plphys/kiab279. Plant Physiol. 2021. PMID: 35237798 Free PMC article.
-
An exceptional horizontal gene transfer in plastids: gene replacement by a distant bacterial paralog and evidence that haptophyte and cryptophyte plastids are sisters.BMC Biol. 2006 Sep 6;4:31. doi: 10.1186/1741-7007-4-31. BMC Biol. 2006. PMID: 16956407 Free PMC article.
-
Massive mitochondrial gene transfer in a parasitic flowering plant clade.PLoS Genet. 2013;9(2):e1003265. doi: 10.1371/journal.pgen.1003265. Epub 2013 Feb 14. PLoS Genet. 2013. PMID: 23459037 Free PMC article.
-
Morphology and phylogenetics of two holoparasitic plants, Balanophora japonica and Balanophora yakushimensis (Balanophoraceae), and their hosts in Taiwan and Japan.J Plant Res. 2012 May;125(3):317-26. doi: 10.1007/s10265-011-0447-5. Epub 2011 Sep 6. J Plant Res. 2012. PMID: 21894574
References
-
- Soltis DE, Soltis PS, Chase MW, Mort ME, Albach DC, Zanis M, Savolainen V, Hahn WH, Hoot SB, Fay MF, Axtell M, Swensen SM, Prince LM, Kress WJ, Nixon KC, Farris JS. Angiosperm phylogeny inferred from 18S rDNA, rbcL, and atpB sequences. Bot Jour Linn Soc. 2000;133:381–461. doi: 10.1006/bojl.2000.0380. - DOI
-
- APG An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Bot Jour Linn Soc. 2003;141:399–436.
-
- Nickrent DL, Duff RJ, Colwell AE, Wolfe AD, Young ND, Steiner KE, dePamphilis CW. Molecular phylogenetic and evolutionary studies of parasitic plants. In: Soltis DE, Soltis PS, Doyle JJ, editor. Molecular Systematics of Plants II. DNA Sequencing. 2. Boston, MA: Kluwer Academic Publishers; 1998. pp. 211–241.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

