Total synthesis of long DNA sequences: synthesis of a contiguous 32-kb polyketide synthase gene cluster
- PMID: 15496466
- PMCID: PMC524854
- DOI: 10.1073/pnas.0406911101
Total synthesis of long DNA sequences: synthesis of a contiguous 32-kb polyketide synthase gene cluster
Abstract
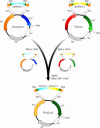
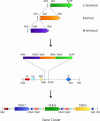
To exploit the huge potential of whole-genome sequence information, the ability to efficiently synthesize long, accurate DNA sequences is becoming increasingly important. An approach proposed toward this end involves the synthesis of approximately 5-kb segments of DNA, followed by their assembly into longer sequences by conventional cloning methods [Smith, H. O., Hutchinson, C. A., III, Pfannkoch, C. & Venter, J. C. (2003) Proc. Natl. Acad. Sci. USA 100, 15440-15445]. The major current impediment to the success of this tactic is the difficulty of building the approximately 5-kb components accurately, efficiently, and rapidly from short synthetic oligonucleotide building blocks. We have developed and implemented a strategy for the high-throughput synthesis of long, accurate DNA sequences. Unpurified 40-base synthetic oligonucleotides are built into 500- to 800-bp "synthons" with low error frequency by automated PCR-based gene synthesis. By parallel processing, these synthons are efficiently joined into multisynthon approximately 5-kb segments by using only three endonucleases and "ligation by selection." These large segments can be subsequently assembled into very long sequences by conventional cloning. We validated the approach by building a synthetic 31,656-bp polyketide synthase gene cluster whose functionality was demonstrated by its ability to produce the megaenzyme and its polyketide product in Escherichia coli.
Figures

References
-
- Khorana, H. G., Yamada, T., Weber, H., Terao, T., RajBhandary, U. L., Otsuka, E., Kumar, A., Gupta, N. K., Buchi, H., Agarwal, K. L., et al. (1972) J. Mol. Biol. 72, 209-217. - PubMed
-
- Sekiya, T., Takeya, T., Brown, E. L., Belagaje, R., Contreras, R., Fritz, H. J., Gait, M. J., Lees, R. G., Ryan, M. J., Khorana, H. G., et al. (1979) J. Biol. Chem. 254, 5787-5801. - PubMed
-
- Itakura, K., Hirose, T., Crea, A. D., Riggs, A. D., Heyneker, H. L., Bolivar, F. & Boyer, H. W. (1977) Science 198, 1056-1063. - PubMed
-
- Dillon, P. J. & Rosen, C. A. (1990) BioTechniques 9, 298-300. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

