A base triple in the Tetrahymena group I core affects the reaction equilibrium via a threshold effect
- PMID: 15496521
- PMCID: PMC1370661
- DOI: 10.1261/rna.7118104
A base triple in the Tetrahymena group I core affects the reaction equilibrium via a threshold effect
Abstract
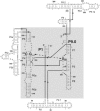
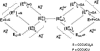
Previous work on group I introns has suggested that a central base triple might be more important for the first rather than the second step of self-splicing, leading to a model in which the base triple undergoes a conformational change during self-splicing. Here, we use the well-characterized L-21 ScaI ribozyme derived from the Tetrahymena group I intron to probe the effects of base-triple disruption on individual reaction steps. Consistent with previous results, reaction of a ternary complex mimicking the first chemical step in self-splicing is slowed by mutations in this base triple, whereas reaction of a ternary complex mimicking the second step of self-splicing is not. Paradoxically, mechanistic dissection of the base-triple disruption mutants indicates that active site binding is weakened uniformly for the 5'-splice site and the 5'-exon analog, mimics for the species bound in the first and second step of self-splicing. Nevertheless, the 5'-exon analog remains bound at the active site, whereas the 5'-splice site analog does not. This differential effect arises despite the uniform destabilization, because the wild-type ribozyme binds the 5'-exon analog more strongly in the active site than in the 5'-splice site analog. Thus, binding into the active site constitutes an additional barrier to reaction of the 5'-splice site analog, but not the 5'-exon analog, resulting in a reduced reaction rate constant for the first step analog, but not the second step analog. This threshold model explains the self-splicing observations without the need to invoke a conformational change involving the base triple, and underscores the importance of quantitative dissection for the interpretation of effects from mutations.
Figures
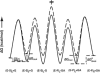

Similar articles
-
Mechanistic investigations of a ribozyme derived from the Tetrahymena group I intron: insights into catalysis and the second step of self-splicing.Biochemistry. 1996 May 7;35(18):5796-809. doi: 10.1021/bi9527653. Biochemistry. 1996. PMID: 8639540
-
A mechanistic framework for the second step of splicing catalyzed by the Tetrahymena ribozyme.Biochemistry. 1996 Jan 16;35(2):648-58. doi: 10.1021/bi951962z. Biochemistry. 1996. PMID: 8555239
-
Self-splicing of the Tetrahymena group I ribozyme without conserved base-triples.Genes Cells. 2001 May;6(5):411-20. doi: 10.1046/j.1365-2443.2001.00437.x. Genes Cells. 2001. PMID: 11380619
-
Ribozymes to the rescue: repairing genetically defective mRNAs.Trends Genet. 1998 Aug;14(8):295-8. doi: 10.1016/s0168-9525(98)01530-3. Trends Genet. 1998. PMID: 9724958 Review. No abstract available.
-
Maximizing RNA folding rates: a balancing act.RNA. 2000 Jun;6(6):790-4. doi: 10.1017/s1355838200000522. RNA. 2000. PMID: 10864039 Free PMC article. Review.
Cited by
-
Alternative substrate kinetics of Escherichia coli ribonuclease P: determination of relative rate constants by internal competition.J Biol Chem. 2013 Mar 22;288(12):8342-8354. doi: 10.1074/jbc.M112.435420. Epub 2013 Jan 28. J Biol Chem. 2013. PMID: 23362254 Free PMC article.
-
Cryo-EM structures of full-length Tetrahymena ribozyme at 3.1 Å resolution.Nature. 2021 Aug;596(7873):603-607. doi: 10.1038/s41586-021-03803-w. Epub 2021 Aug 11. Nature. 2021. PMID: 34381213 Free PMC article.
-
A kinetic and thermodynamic framework for the Azoarcus group I ribozyme reaction.RNA. 2014 Nov;20(11):1732-46. doi: 10.1261/rna.044362.114. Epub 2014 Sep 22. RNA. 2014. PMID: 25246656 Free PMC article.
-
PseudoViewer: web application and web service for visualizing RNA pseudoknots and secondary structures.Nucleic Acids Res. 2006 Jul 1;34(Web Server issue):W416-22. doi: 10.1093/nar/gkl210. Nucleic Acids Res. 2006. PMID: 16845039 Free PMC article.
-
How to Kinetically Dissect an RNA Machine.Biochemistry. 2021 Nov 23;60(46):3485-3490. doi: 10.1021/acs.biochem.1c00392. Epub 2021 Sep 7. Biochemistry. 2021. PMID: 34492193 Free PMC article.
References
-
- Adams, P.L., Stahley, M.R., Kosek, A.B., Wang, J., and Strobel, S.A. 2004. Crystal structure of a self-splicing group I intron with both exons. Nature 430: 45–50. - PubMed
-
- Bartley, L.E., Zhuang, X., Das, R., Chu, S., and Herschlag, D. 2003. Exploration of the transition state for tertiary structure formation between an RNA helix and a large structured RNA. J. Mol. Biol. 328: 1011–1026. - PubMed
-
- Bevilacqua, P.C., Kierzek, R., Johnson, K.A., and Turner, D.H. 1992. Dynamics of ribozyme binding of substrate revealed by fluorescence-detected stopped-flow methods. Science 258: 1355–1357. - PubMed
-
- Bevilacqua, P.C., Li, Y., and Turner, D.H. 1994. Fluorescence-detected stopped-flow With a pyrene-labeled substrate reveals that guano-sine facilitates docking of the 5′ cleavage site into a high free-energy binding mode in the Tetrahymena ribozyme. Biochemistry 33: 11340–11348. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
