The PRC-barrel domain of the ribosome maturation protein RimM mediates binding to ribosomal protein S19 in the 30S ribosomal subunits
- PMID: 15496525
- PMCID: PMC1370667
- DOI: 10.1261/rna.7720204
The PRC-barrel domain of the ribosome maturation protein RimM mediates binding to ribosomal protein S19 in the 30S ribosomal subunits
Abstract
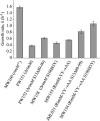
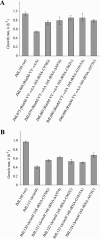
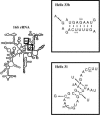
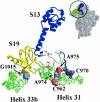
The RimM protein in Escherichia coli is associated with free 30S ribosomal subunits but not with 70S ribosomes. A DeltarimM mutant is defective in 30S maturation and accumulates 17S rRNA. To study the interaction of RimM with the 30S and its involvement in 30S maturation, RimM amino acid substitution mutants were constructed. A mutant RimM (RimM-YY-->AA), containing alanine substitutions for two adjacent tyrosines within the PRC beta-barrel domain, showed a reduced binding to 30S and an accumulation of 17S rRNA compared to wild-type RimM. The (RimM-YY-->AA) and DeltarimM mutants had significantly lower amounts of polysomes and also reduced levels of 30S relative to 50S compared to a wild-type strain. A mutation in rpsS, which encodes r-protein S19, suppressed the polysome- and 16S rRNA processing deficiencies of the RimM-YY-->AA but not that of the DeltarimM mutant. A mutation in rpsM, which encodes r-protein S13, suppressed the polysome deficiency of both rimM mutants. Suppressor mutations, found in either helices 31 or 33b of 16S rRNA, improved growth of both the RimM-YY-->AA and DeltarimM mutants. However, they suppressed the 16S rRNA processing deficiency of the RimM-YY-->AA mutant more efficiently than that of the DeltarimM mutant. Helices 31 and 33b are known to interact with S13 and S19, respectively, and S13 is known to interact with S19. A GST-RimM but not a GST-RimM(YY-->AA) protein bound strongly to S19 in 30S. Thus, RimM likely facilitates maturation of the region of the head of 30S that contains S13 and S19 as well as helices 31 and 33b.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
