A functional role of postsynaptic density-95-guanylate kinase-associated protein complex in regulating Shank assembly and stability to synapses
- PMID: 15496675
- PMCID: PMC6730104
- DOI: 10.1523/JNEUROSCI.3314-04.2004
A functional role of postsynaptic density-95-guanylate kinase-associated protein complex in regulating Shank assembly and stability to synapses
Abstract
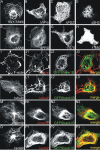
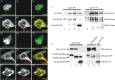
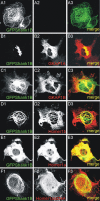
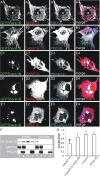
Postsynaptic density (PSD) proteins include scaffold, cytoskeletal, and signaling proteins that structurally and functionally interact with glutamate receptors and other postsynaptic membrane proteins. The molecular mechanisms regulating the assembly of PSD proteins and their associations with synapses are still widely unknown. We investigated the molecular mechanisms of Shank1 targeting and synapse assembly by looking at the function of guanylate kinase-associated protein (GKAP) and PSD-95 interactions. Shank1 when it is not associated to GKAP, which binds to the Shank PSD-95-Discs Large-zona occludens-1 domain, forms filamentous and fusiform structures in which the Src homology 3 domain specifically interacts with the ankyrin repeat domain, thus allowing its multimerization via a novel form of intermolecular interaction. Surprisingly, in both COS-7 cells and hippocampal neurons, GKAP forms insoluble aggregates with Shank that colocalize with heat shock protein 70 and neurofilaments, two markers of the aggresomes in which misfolded proteins accumulate. However, the two proteins are organized in clusters in COS cells and synaptic clusters in neurons when both are overexpressed and associated with wild-type PSD-95, but not with palmitoylation-deficient PSD-95. Synaptic activity in neurons induces the formation of Shank and GKAP intracellular aggregation and degradation. Similarly, the overexpression of a GKAP mutant that is incapable of binding PSD-95 induces Shank aggregation and degradation in neurons. Our data suggest a possible functional and structural role of the PSD-95-GKAP complex in Shank and PSD protein assembly and stability to synapses.
Figures






References
-
- Beneken J, Tu JC, Xiao B, Nuriya M, Yuan JP, Worley PF, Leahy DJ (2000) Structure of the Homer EVH1 domain-peptide complex reveals a new twist in polyproline recognition. Neuron 26: 143-154. - PubMed
-
- Bockmann J, Kreutz MR, Gundelfinger ED, Bockers TM (2002) ProSAP/Shank postsynaptic density proteins interact with insulin receptor tyrosine kinase substrate IRSp53. J Neurochem 83: 1013-1017. - PubMed
-
- Boeckers TM, winter C, Smalla KH, Kreutz MR, Bockmann J, Seidenbecher C, Garner CC, Gundelfinger ED (1999) Proline-rich synapse-associated proteins ProSAP1 and ProSAP2 interact with synaptic proteins of the SAPAP/GKAP family. Biochem Biophys Res Commun 264: 247-252. - PubMed
-
- Boeckers TM, Bockmann J, Kreutz MR, Gundelfinger ED (2002) ProSAP/Shank proteins - a family of higher order organizing molecules of the postsynaptic density with an emerging role in human neurological disease. J Neurochem 81: 903-910. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
