Characterization of phylogenetically distant members of the adenylate cyclase family from mycobacteria: Rv1647 from Mycobacterium tuberculosis and its orthologue ML1399 from M. leprae
- PMID: 15500449
- PMCID: PMC1134983
- DOI: 10.1042/BJ20041040
Characterization of phylogenetically distant members of the adenylate cyclase family from mycobacteria: Rv1647 from Mycobacterium tuberculosis and its orthologue ML1399 from M. leprae
Abstract
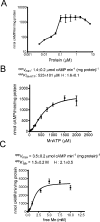
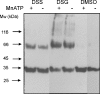
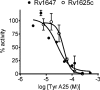
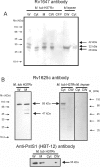
Analysis of the genome sequence of Mycobacterium tuberculosis H37Rv has identified 16 genes that are similar to the mammalian adenylate and guanylate cyclases. Rv1647 was predicted to be an active adenylate cyclase but its position in a phylogenetically distant branch from the other enzymes characterized so far from M. tuberculosis makes it an interestingly divergent nucleotide cyclase to study. In agreement with its divergence at the sequence level from other nucleotide cyclases, the cloning, expression and purification of Rv1647 revealed differences in its biochemical properties from the previously characterized Rv1625c adenylate cyclase. Adenylate cyclase activity of Rv1647 was activated by detergents but was resistant to high concentrations of salt. Mutations of substrate-specifying residues to those present in guanylate cyclases failed to convert the enzyme into a guanylate cyclase, and did not alter its oligomeric status. Orthologues of Rv1647 could be found in M. leprae, M. avium and M. smegmatis. The orthologue from M. leprae (ML1399) was cloned, and the protein was expressed, purified and shown biochemically to be an adenylate cyclase, thus representing the first adenylate cyclase to be described from M. leprae. Importantly, Western-blot analysis of subcellular fractions from M. tuberculosis and M. leprae revealed that the Rv1647 and ML1399 gene products respectively were expressed in these bacteria. Additionally, M. tuberculosis was also found to express the Rv1625c adenylate cyclase, suggesting that multiple adenylate cyclase proteins may be expressed simultaneously in this organism. These results suggest that class III cyclase-like gene products probably have an important role to play in the physiology and perhaps the pathology of these medically important bacteria.
Figures





Similar articles
-
Cyclic AMP in mycobacteria: characterization and functional role of the Rv1647 ortholog in Mycobacterium smegmatis.J Bacteriol. 2008 Jun;190(11):3824-34. doi: 10.1128/JB.00138-08. Epub 2008 Apr 4. J Bacteriol. 2008. PMID: 18390660 Free PMC article.
-
Mutational analysis of the Mycobacterium tuberculosis Rv1625c adenylyl cyclase: residues that confer nucleotide specificity contribute to dimerization.FEBS Lett. 2003 Jun 19;545(2-3):253-9. doi: 10.1016/s0014-5793(03)00580-5. FEBS Lett. 2003. PMID: 12804785
-
An adenylyl cyclase pseudogene in Mycobacterium tuberculosis has a functional ortholog in Mycobacterium avium.Biochimie. 2005 Jun;87(6):557-63. doi: 10.1016/j.biochi.2005.01.017. Biochimie. 2005. PMID: 15908099
-
The class III adenylyl cyclases: multi-purpose signalling modules.Cell Signal. 2003 Dec;15(12):1081-9. doi: 10.1016/s0898-6568(03)00130-x. Cell Signal. 2003. PMID: 14575863 Review.
-
In search of a function for the membrane anchors of class IIIa adenylate cyclases.Int J Med Microbiol. 2019 May-Jun;309(3-4):245-251. doi: 10.1016/j.ijmm.2019.03.006. Epub 2019 Mar 22. Int J Med Microbiol. 2019. PMID: 30954381 Review.
Cited by
-
Characterization of Mycobacterium tuberculosis Rv3676 (CRPMt), a cyclic AMP receptor protein-like DNA binding protein.J Bacteriol. 2005 Nov;187(22):7795-804. doi: 10.1128/JB.187.22.7795-7804.2005. J Bacteriol. 2005. PMID: 16267303 Free PMC article.
-
Cyclic AMP in mycobacteria: characterization and functional role of the Rv1647 ortholog in Mycobacterium smegmatis.J Bacteriol. 2008 Jun;190(11):3824-34. doi: 10.1128/JB.00138-08. Epub 2008 Apr 4. J Bacteriol. 2008. PMID: 18390660 Free PMC article.
-
Overexpression of the Rv0805 phosphodiesterase elicits a cAMP-independent transcriptional response.Tuberculosis (Edinb). 2013 Sep;93(5):492-500. doi: 10.1016/j.tube.2013.05.004. Epub 2013 Jul 6. Tuberculosis (Edinb). 2013. PMID: 23835087 Free PMC article.
-
Mycobacterial STAND adenylyl cyclases: The HTH domain binds DNA to form biocrystallized nucleoids.Biophys J. 2021 Apr 6;120(7):1231-1246. doi: 10.1016/j.bpj.2020.11.008. Epub 2020 Nov 18. Biophys J. 2021. PMID: 33217386 Free PMC article.
-
Profound asymmetry in the structure of the cAMP-free cAMP Receptor Protein (CRP) from Mycobacterium tuberculosis.J Biol Chem. 2009 Mar 27;284(13):8228-32. doi: 10.1074/jbc.C800215200. Epub 2009 Feb 4. J Biol Chem. 2009. PMID: 19193643 Free PMC article.
References
-
- Shenoy A. R., Visweswariah S. S. Class III nucleotide cyclases in bacteria and archaebacteria: lineage-specific expansion of adenylyl cyclases and a dearth of guanylyl cyclases. FEBS Lett. 2004;561:11–21. - PubMed
-
- Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C. E., III, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature (London) 1998;393:537–544. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

