Expression of type IV pili by Moraxella catarrhalis is essential for natural competence and is affected by iron limitation
- PMID: 15501752
- PMCID: PMC523052
- DOI: 10.1128/IAI.72.11.6262-6270.2004
Expression of type IV pili by Moraxella catarrhalis is essential for natural competence and is affected by iron limitation
Abstract
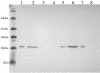
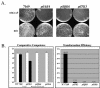
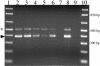
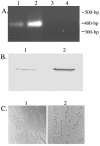
Type IV pili, filamentous surface appendages primarily composed of a single protein subunit termed pilin, play a crucial role in the initiation of disease by a wide range of pathogenic bacteria. Although previous electron microscopic studies suggested that pili might be present on the surface of Moraxella catarrhalis isolates, detailed molecular and phenotypic analyses of these structures have not been reported to date. We identified and cloned the M. catarrhalis genes encoding PilA, the major pilin subunit, PilQ, the outer membrane secretin through which the pilus filament is extruded, and PilT, the NTPase that mediates pilin disassembly and retraction. To initiate investigation of the role of this surface organelle in pathogenesis, isogenic pilA, pilT, and pilQ mutants were constructed in M. catarrhalis strain 7169. Comparative analyses of the wild-type 7169 strain and three isogenic pil mutants demonstrated that M. catarrhalis expresses type IV pili that are essential for natural genetic transformation. Our studies suggest type IV pilus production by M. catarrhalis is constitutive and ubiquitous, although pilin expression was demonstrated to be iron responsive and Fur regulated. These data indicate that additional studies aimed at elucidating the prevalence and role of type IV pili in the pathogenesis and host response to M. catarrhalis infections are warranted.
Figures


References
-
- Aas, F. E., M. Wolfgang, S. Frye, S. Dunham, C. Lovold, and M. Koomey. 2002. Competence for natural transformation in Neisseria gonorrhoeae: components of DNA binding and uptake linked to type IV pilus expression. Mol. Microbiol. 46:749-760. - PubMed
-
- Ahmed, K. 1992. Fimbriae of Branhamella catarrhalis as possible mediators of adherence to pharyngeal epithelial cells. APMIS 100:1066-1072. - PubMed
-
- Ahmed, K., N. Rikitomi, and K. Matsumoto. 1992. Fimbriation, hemagglutination and adherence properties of fresh clinical isolates of Branhamella catarrhalis. Microbiol. Immunol. 36:1009-1017. - PubMed
-
- Ahmed, K., N. Rikitomi, T. Nagatake, and K. Matsumoto. 1990. Electron microscopic observation of Branhamella catarrhalis. Microbiol. Immunol. 34:967-975. - PubMed
-
- Alm, R. A., and J. S. Mattick. 1997. Genes involved in the biogenesis and function of type-4 fimbriae in Pseudomonas aeruginosa. Gene 192:89-98. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

