Crystal structure of the dimeric protein core of decorin, the archetypal small leucine-rich repeat proteoglycan
- PMID: 15501918
- PMCID: PMC524833
- DOI: 10.1073/pnas.0402976101
Crystal structure of the dimeric protein core of decorin, the archetypal small leucine-rich repeat proteoglycan
Abstract
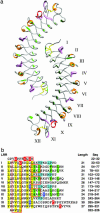
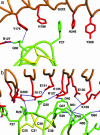
Decorin is a ubiquitous extracellular matrix proteoglycan with a variety of important biological functions that are mediated by its interactions with extracellular matrix proteins, cytokines, and cell surface receptors. Decorin is the prototype of the family of small leucine-rich repeat proteoglycans and proteins (SLRPs), characterized by a protein core composed of leucine-rich repeats (LRRs), flanked by two cysteine-rich regions. We report here the crystal structure of the dimeric protein core of decorin, the best characterized member of the SLRP family. Each monomer adopts the curved solenoid fold characteristic of LRR domains, with a parallel beta-sheet on the inside interwoven with loops containing short segments of beta-strands, 3(10) helices, and polyproline II helices on the outside. Two main features are unique to this structure. First, decorin dimerizes through the concave surfaces of the LRR domains, which have been implicated previously in protein-ligand interactions. The amount of surface buried in this dimer rivals the buried surfaces of some of the highest-affinity macromolecular complexes reported to date. Second, the C-terminal region adopts an unusual capping motif that involves a laterally extended LRR and a disulfide bond. This motif seems to be unique to SLRPs and has not been observed in any other LRR protein structure to date. Possible implications of these features for decorin ligand binding and SLRP function are discussed.
Figures


Similar articles
-
Structural correlations in the family of small leucine-rich repeat proteins and proteoglycans.J Struct Biol. 2006 Aug;155(2):294-305. doi: 10.1016/j.jsb.2006.01.016. Epub 2006 May 19. J Struct Biol. 2006. PMID: 16884925 Review.
-
LRRCE: a leucine-rich repeat cysteine capping motif unique to the chordate lineage.BMC Genomics. 2008 Dec 12;9:599. doi: 10.1186/1471-2164-9-599. BMC Genomics. 2008. PMID: 19077264 Free PMC article.
-
Crystal structure of the biglycan dimer and evidence that dimerization is essential for folding and stability of class I small leucine-rich repeat proteoglycans.J Biol Chem. 2006 May 12;281(19):13324-13332. doi: 10.1074/jbc.M513470200. Epub 2006 Mar 17. J Biol Chem. 2006. PMID: 16547006
-
Leucine-rich repeat region of decorin binds to filamin-A.Biochimie. 2002 Apr;84(4):303-8. doi: 10.1016/s0300-9084(02)01391-3. Biochimie. 2002. PMID: 12106908
-
Leucine-rich repeat glycoproteins of the extracellular matrix.Matrix Biol. 1998 Apr;17(1):1-19. doi: 10.1016/s0945-053x(98)90121-4. Matrix Biol. 1998. PMID: 9628249 Review.
Cited by
-
Differential availability/processing of decorin precursor in arterial and venous smooth muscle cells.J Anat. 2006 Sep;209(3):271-87. doi: 10.1111/j.1469-7580.2006.00614.x. J Anat. 2006. PMID: 16928198 Free PMC article.
-
Generation of a multi-functional, target organ-specific, anti-fibrotic molecule by molecular engineering of the extracellular matrix protein, decorin.Br J Pharmacol. 2019 Jan;176(1):16-25. doi: 10.1111/bph.14374. Epub 2018 Jun 25. Br J Pharmacol. 2019. PMID: 29847688 Free PMC article. Review.
-
Decorin interacts with connective tissue growth factor (CTGF)/CCN2 by LRR12 inhibiting its biological activity.J Biol Chem. 2011 Jul 8;286(27):24242-52. doi: 10.1074/jbc.M110.189365. Epub 2011 Mar 23. J Biol Chem. 2011. PMID: 21454550 Free PMC article.
-
Comparative Geometrical Analysis of Leucine-Rich Repeat Structures in the Nod-Like and Toll-Like Receptors in Vertebrate Innate Immunity.Biomolecules. 2015 Aug 18;5(3):1955-78. doi: 10.3390/biom5031955. Biomolecules. 2015. PMID: 26295267 Free PMC article. Review.
-
The C-terminal peptide of chondroadherin modulates cellular activity by selectively binding to heparan sulfate chains.J Biol Chem. 2013 Jan 11;288(2):995-1008. doi: 10.1074/jbc.M112.430512. Epub 2012 Nov 21. J Biol Chem. 2013. PMID: 23172228 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

