Unidirectional translocation from recognition site and a necessary interaction with DNA end for cleavage by Type III restriction enzyme
- PMID: 15501920
- PMCID: PMC528788
- DOI: 10.1093/nar/gkh899
Unidirectional translocation from recognition site and a necessary interaction with DNA end for cleavage by Type III restriction enzyme
Abstract
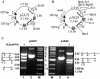
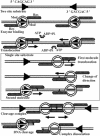
Type III restriction enzymes have been demonstrated to require two unmethylated asymmetric recognition sites oriented head-to-head to elicit double-strand break 25-27 bp downstream of one of the two sites. The proposed DNA cleavage mechanism involves ATP-dependent DNA translocation. The sequence context of the recognition site was suggested to influence the site of DNA cleavage by the enzyme. In this investigation, we demonstrate that the cleavage site of the R.EcoP15I restriction enzyme does not depend on the sequence context of the recognition site. Strikingly, this study demonstrates that the enzyme can cleave linear DNA having either recognition sites in the same orientation or a single recognition site. Cleavage occurs predominantly at a site proximal to the DNA end in the case of multiple site substrates. Such cleavage can be abolished by the binding of Lac repressor downstream (3' side) but not upstream (5' side) of the recognition site. Binding of HU protein has also been observed to interfere with R.EcoP15I cleavage activity. In accordance with a mechanism requiring two enzyme molecules cooperating to elicit double-strand break on DNA, our results convincingly demonstrate that the enzyme translocates on DNA in a 5' to 3' direction from its recognition site and indicate a switch in the direction of enzyme motion at the DNA ends. This study demonstrates a new facet in the mode of action of these restriction enzymes.
Figures







Similar articles
-
DNA cleavage by type III restriction-modification enzyme EcoP15I is independent of spacer distance between two head to head oriented recognition sites.J Mol Biol. 2001 Sep 28;312(4):687-98. doi: 10.1006/jmbi.2001.4998. J Mol Biol. 2001. PMID: 11575924
-
Subunit assembly and mode of DNA cleavage of the type III restriction endonucleases EcoP1I and EcoP15I.J Mol Biol. 2001 Feb 23;306(3):417-31. doi: 10.1006/jmbi.2000.4411. J Mol Biol. 2001. PMID: 11178902
-
Scanning force microscopy of DNA translocation by the Type III restriction enzyme EcoP15I.J Mol Biol. 2004 Aug 6;341(2):337-43. doi: 10.1016/j.jmb.2004.06.031. J Mol Biol. 2004. PMID: 15276827
-
How to get from A to B: strategies for analysing protein motion on DNA.Eur Biophys J. 2002 Jul;31(4):257-67. doi: 10.1007/s00249-002-0224-4. Epub 2002 May 30. Eur Biophys J. 2002. PMID: 12122472 Review.
-
Does topoisomerase II specifically recognize and cleave hairpins, cruciforms and crossovers of DNA?Biochimie. 2007 Apr;89(4):508-15. doi: 10.1016/j.biochi.2007.02.011. Epub 2007 Feb 24. Biochimie. 2007. PMID: 17397986 Review.
Cited by
-
Dissociation from DNA of Type III Restriction-Modification enzymes during helicase-dependent motion and following endonuclease activity.Nucleic Acids Res. 2012 Aug;40(14):6752-64. doi: 10.1093/nar/gks328. Epub 2012 Apr 20. Nucleic Acids Res. 2012. PMID: 22523084 Free PMC article.
-
Translocation, switching and gating: potential roles for ATP in long-range communication on DNA by Type III restriction endonucleases.Biochem Soc Trans. 2011 Apr;39(2):589-94. doi: 10.1042/BST0390589. Biochem Soc Trans. 2011. PMID: 21428945 Free PMC article. Review.
-
Type III restriction-modification enzymes: a historical perspective.Nucleic Acids Res. 2014 Jan;42(1):45-55. doi: 10.1093/nar/gkt616. Epub 2013 Jul 17. Nucleic Acids Res. 2014. PMID: 23863841 Free PMC article. Review.
-
Conflicts targeting epigenetic systems and their resolution by cell death: novel concepts for methyl-specific and other restriction systems.DNA Res. 2010 Dec;17(6):325-42. doi: 10.1093/dnares/dsq027. Epub 2010 Nov 8. DNA Res. 2010. PMID: 21059708 Free PMC article. Review.
-
The single polypeptide restriction-modification enzyme LlaGI is a self-contained molecular motor that translocates DNA loops.Nucleic Acids Res. 2009 Nov;37(21):7219-30. doi: 10.1093/nar/gkp794. Nucleic Acids Res. 2009. PMID: 19783815 Free PMC article.
References
-
- Gorbalenya A.E. and Koonin,E.V. (1991) Endonuclease (R) subunits of type-I and type-III restriction-modification enzymes contain a helicase-like domain. FEBS Lett., 291, 277–281. - PubMed
-
- Rao D.N., Saha,S. and Krishnamurthy,V. (2000) ATP-dependent restriction enzymes. Prog. Nucleic Acid Res. Mol. Biol., 64, 1–63. - PubMed
-
- Reiser J. and Yuan,R. (1977) Purification and properties of the P15-specific restriction endonuclease from Escherichia coli. J. Biol. Chem., 252, 451–456. - PubMed
-
- Hadi S.M., Bächi,B., Shepherd,J.C., Yuan,R., Ineichen,K. and Bickle,T.A. (1979) DNA recognition and cleavage by the EcoP15 restriction endonuclease. J. Mol. Biol., 134, 655–666. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

