Expression, purification, and biochemical characterization of the antiinflammatory tristetraprolin: a zinc-dependent mRNA binding protein affected by posttranslational modifications
- PMID: 15504035
- PMCID: PMC1351390
- DOI: 10.1021/bi049014y
Expression, purification, and biochemical characterization of the antiinflammatory tristetraprolin: a zinc-dependent mRNA binding protein affected by posttranslational modifications
Abstract
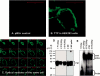
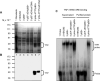
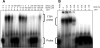
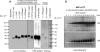
Tristetraprolin (TTP) is a hyperphosphorylated protein that destabilizes mRNA by binding to an AU-rich element (ARE). Mice deficient in TTP develop a severe inflammatory syndrome. The biochemical properties of TTP have not been adequately characterized, due to the difficulties in protein purification and lack of a high-titer antiserum. Full-length human TTP was expressed in human HEK293 cells and purified to at least 70% homogeneity. The purified protein was free of endogenous ARE binding activity, and was used for investigating its size, zinc dependency, and binding kinetics for tumor necrosis factor alpha mRNA ARE. A high-titer rabbit antiserum was raised against the MBP-hTTP fusion protein expressed in Escherichia coli. Cellular localization studies of the transfected cells indicated that approximately 80% of the expressed TTP was in the cytosol, with 20% in the nuclei. TTP from both locations bound to the ARE and formed similar complexes. The purified TTP was shown to be intact by N-terminal His-tag purification, C-terminal peptide sequencing, and mass spectrometry analysis. Results from size exclusion chromatography are consistent with the predominant form of active TTP being a tetramer. TTP's ARE binding activity was increased by 10 microM Zn(2+). The half-maximal binding of TTP from HEK293 cells was approximately 30 nM in assays containing 10 nM ARE. This value was about twice that of TTP from E. coli. TTP from HEK293 cells was highly phosphorylated, and its electrophoretic mobility was increased by alkaline phosphatase treatment and somewhat by T271A mutation, but not by PNGase F or S186A mutation. The gel mobility of TTP from E. coli was decreased by in vitro phosphorylation with p42/ERK2 and p38 mitogen-activated protein kinases. These results suggest that TTP's zinc-dependent ARE binding affinity is reduced by half by posttranslational modifications, mainly by phosphorylation but not by glycosylation, in mammalian cells. The results support a model in which each subunit of the TTP tetramer binds to one of the five overlapping UUAUUUAUU sequences of the ARE, resulting in a stable TTP-ARE complex.
Figures




References
-
- Blackshear PJ. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem. Soc. Trans. 2002;30:945–952. - PubMed
-
- Lai WS, Stumpo DJ, Blackshear PJ. Rapid insulin-stimulated accumulation of an mRNA encoding a proline-rich protein. J. Biol. Chem. 1990;265:16556–16563. - PubMed
-
- Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science. 1998;281:1001–1005. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

